The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Fiedorczuk, K., Iordanov, I., Mihalyi, C., Szollosi, A., Csanady, L., Chen, J.(2024) Proc Natl Acad Sci U S A 121: e2409049121-e2409049121
- PubMed: 39495916
- DOI: https://doi.org/10.1073/pnas.2409049121
- Primary Citation of Related Structures:
9DW4, 9DW5, 9DW7, 9DW8, 9DW9 - PubMed Abstract:
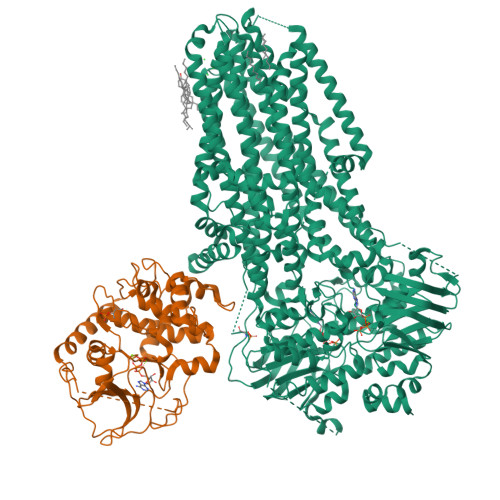
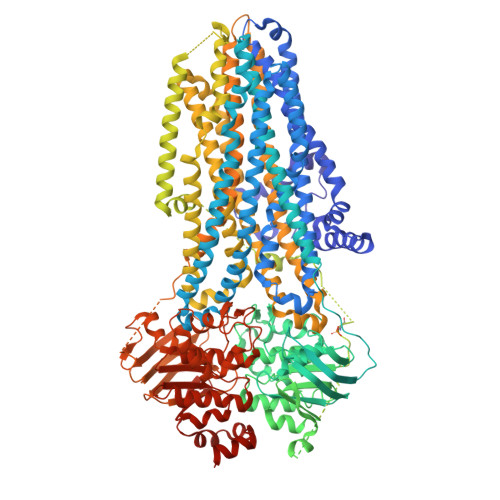
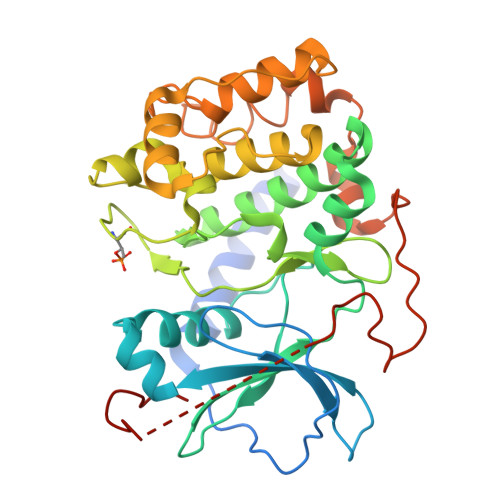
Protein kinase A (PKA) is a key regulator of cellular functions by selectively phosphorylating numerous substrates, including ion channels, enzymes, and transcription factors. It has long served as a model system for understanding the eukaryotic kinases. Using cryoelectron microscopy, we present complex structures of the PKA catalytic subunit (PKA-C) bound to a full-length protein substrate, the cystic fibrosis transmembrane conductance regulator (CFTR)-an ion channel vital to human health. CFTR gating requires phosphorylation of its regulatory (R) domain. Unphosphorylated CFTR engages PKA-C at two locations, establishing two "catalytic stations" near to, but not directly involving, the R domain. This configuration, coupled with the conformational flexibility of the R domain, permits transient interactions of the eleven spatially separated phosphorylation sites. Furthermore, we determined two structures of the open-pore CFTR stabilized by PKA-C, providing a molecular basis to understand how PKA-C stimulates CFTR currents even in the absence of phosphorylation.
Organizational Affiliation:
Laboratory of Membrane Biology and Biophysics, The Rockefeller University, New York, NY 10065.