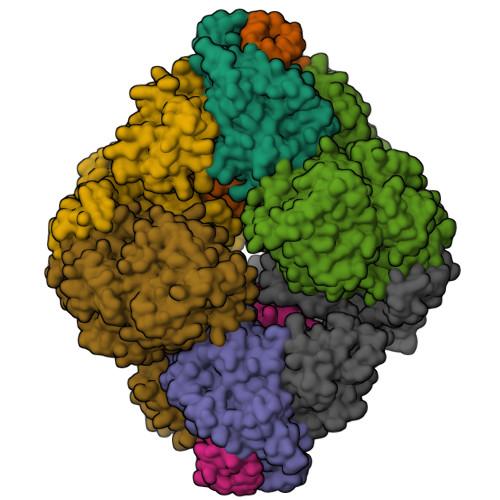
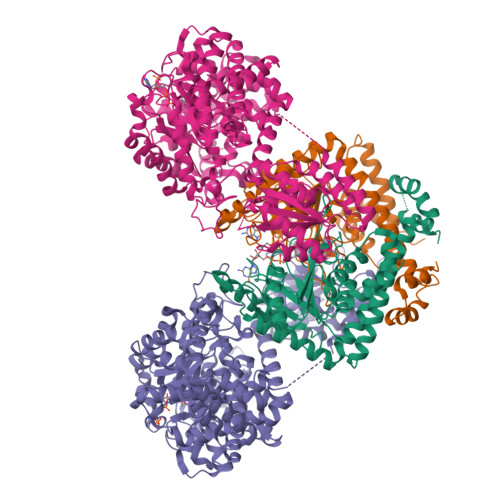
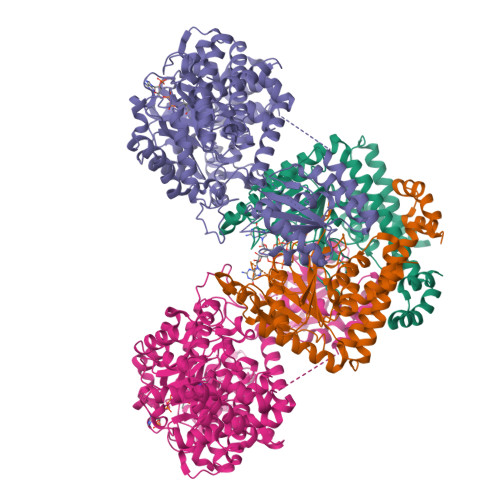
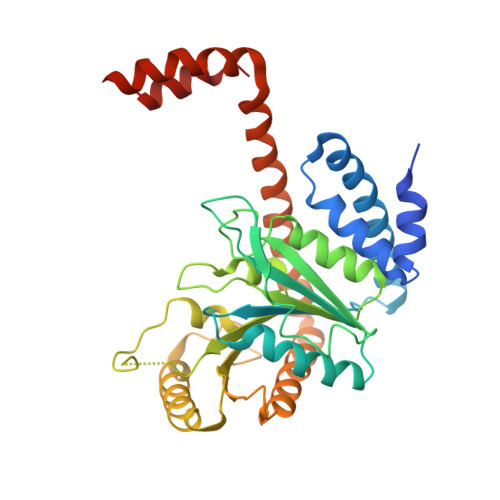
Architecture of the human G-protein-methylmalonyl-CoA mutase nanoassembly for B 12 delivery and repair.
Mascarenhas, R., Ruetz, M., Gouda, H., Heitman, N., Yaw, M., Banerjee, R.(2023) Nat Commun 14: 4332-4332
- PubMed: 37468522
- DOI: https://doi.org/10.1038/s41467-023-40077-4
- Primary Citation of Related Structures:
8GJU - PubMed Abstract:
G-proteins function as molecular switches to power cofactor translocation and confer fidelity in metal trafficking. The G-protein, MMAA, together with MMAB, an adenosyltransferase, orchestrate cofactor delivery and repair of B 12 -dependent human methylmalonyl-CoA mutase (MMUT). The mechanism by which the complex assembles and moves a >1300 Da cargo, or fails in disease, are poorly understood. Herein, we report the crystal structure of the human MMUT-MMAA nano-assembly, which reveals a dramatic 180° rotation of the B 12 domain, exposing it to solvent. The complex, stabilized by MMAA wedging between two MMUT domains, leads to ordering of the switch I and III loops, revealing the molecular basis of mutase-dependent GTPase activation. The structure explains the biochemical penalties incurred by methylmalonic aciduria-causing mutations that reside at the MMAA-MMUT interfaces we identify here.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, MI, 48109, USA.