Evolution-inspired engineering of anthracycline methyltransferases.
Dinis, P., Tirkkonen, H., Wandi, B.N., Siitonen, V., Niemi, J., Grocholski, T., Metsa-Ketela, M.(2023) PNAS Nexus 2: pgad009-pgad009
- PubMed: 36874276
- DOI: https://doi.org/10.1093/pnasnexus/pgad009
- Primary Citation of Related Structures:
7OWB, 7OY1, 7PG7, 7PGA, 7PGJ, 7PHD, 7PHE, 7PHF - PubMed Abstract:
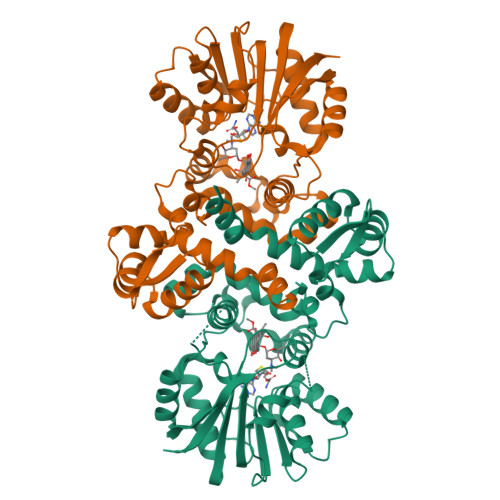
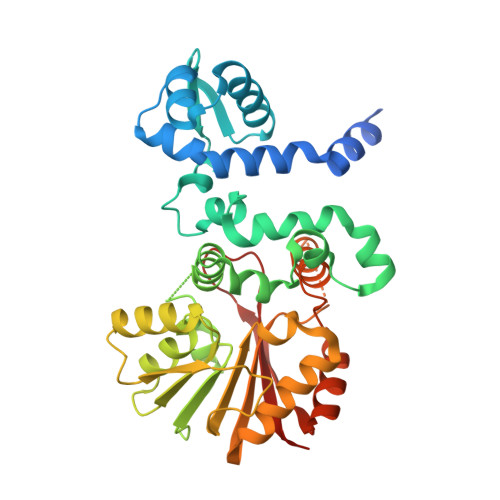
Streptomyces soil bacteria produce hundreds of anthracycline anticancer agents with a relatively conserved set of genes. This diversity depends on the rapid evolution of biosynthetic enzymes to acquire novel functionalities. Previous work has identified S -adenosyl-l-methionine-dependent methyltransferase-like proteins that catalyze 4-O-methylation, 10-decarboxylation, or 10-hydroxylation, with additional differences in substrate specificities. Here we focused on four protein regions to generate chimeric enzymes using sequences from four distinct subfamilies to elucidate their influence in catalysis. Combined with structural studies we managed to depict factors that influence gain-of-hydroxylation, loss-of-methylation, and substrate selection. The engineering expanded the catalytic repertoire to include novel 9,10-elimination activity, and 4-O-methylation and 10-decarboxylation of unnatural substrates. The work provides an instructive account on how the rise of diversity of microbial natural products may occur through subtle changes in biosynthetic enzymes.
Organizational Affiliation:
Department of Life Technologies, University of Turku, BioCity, Tykistökatu 6, FIN-20014 Turku, Finland.