Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent.
Kurasawa, O., Miyazaki, T., Homma, M., Oguro, Y., Imada, T., Uchiyama, N., Iwai, K., Yamamoto, Y., Ohori, M., Hara, H., Sugimoto, H., Iwata, K., Skene, R., Hoffman, I., Ohashi, A., Nomura, T., Cho, N.(2020) J Med Chem 63: 1084-1104
- PubMed: 31895562
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01427
- Primary Citation of Related Structures:
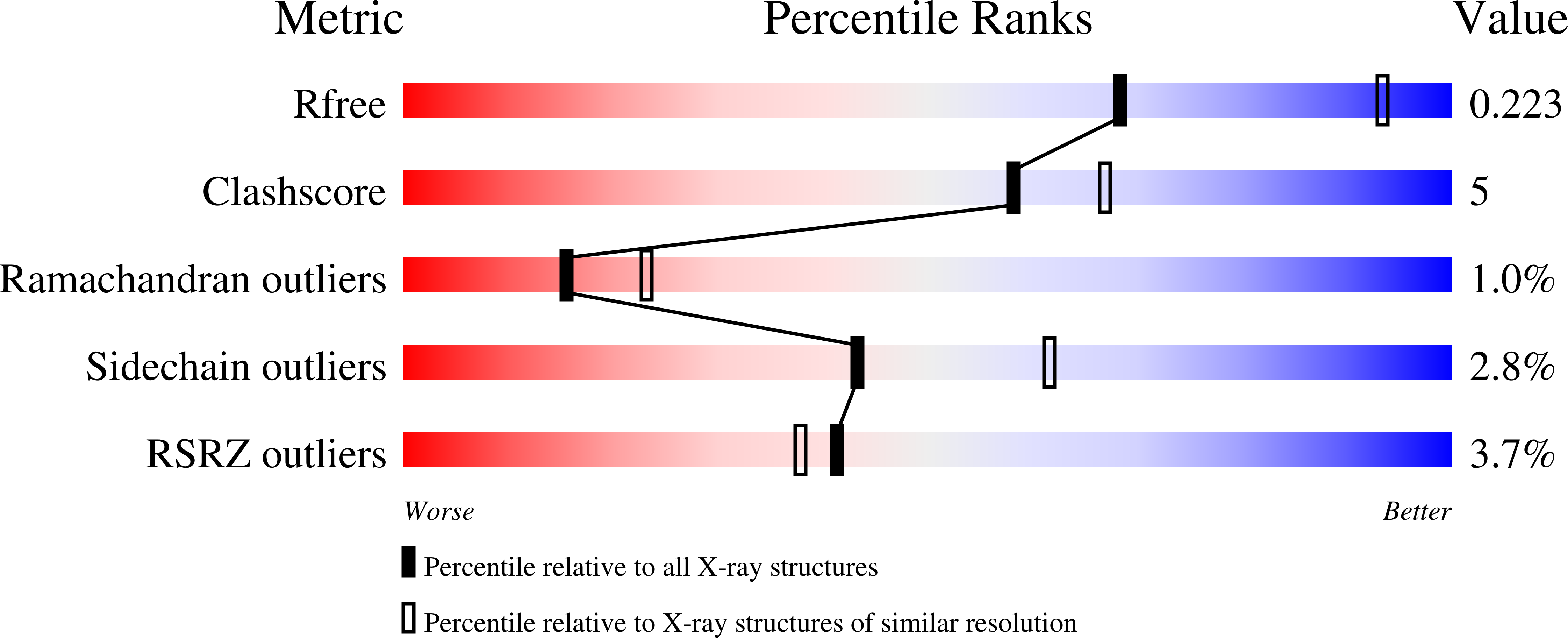
6P5M, 6P5P - PubMed Abstract:
In our pursuit of developing a novel, potent, and selective cell division cycle 7 (Cdc7) inhibitor, we optimized the previously reported thieno[3,2- d ]pyrimidinone analogue I showing time-dependent Cdc7 kinase inhibition and slow dissociation kinetics. These medicinal chemistry efforts led to the identification of compound 3d , which exhibited potent cellular activity, excellent kinase selectivity, and antitumor efficacy in a COLO205 xenograft mouse model. However, the issue of formaldehyde adduct formation emerged during a detailed study of 3d , which was deemed an obstacle to further development. A structure-based approach to circumvent the adduct formation culminated in the discovery of compound 11b ( TAK-931 ) possessing a quinuclidine moiety as a preclinical candidate. In this paper, the design, synthesis, and biological evaluation of this series of compounds will be presented.
Organizational Affiliation:
Pharmaceutical Research Division , Takeda Pharmaceutical Company, Ltd. , 26-1, Muraoka-Higashi 2-chome , Fujisawa , Kanagawa 251-8555 , Japan.