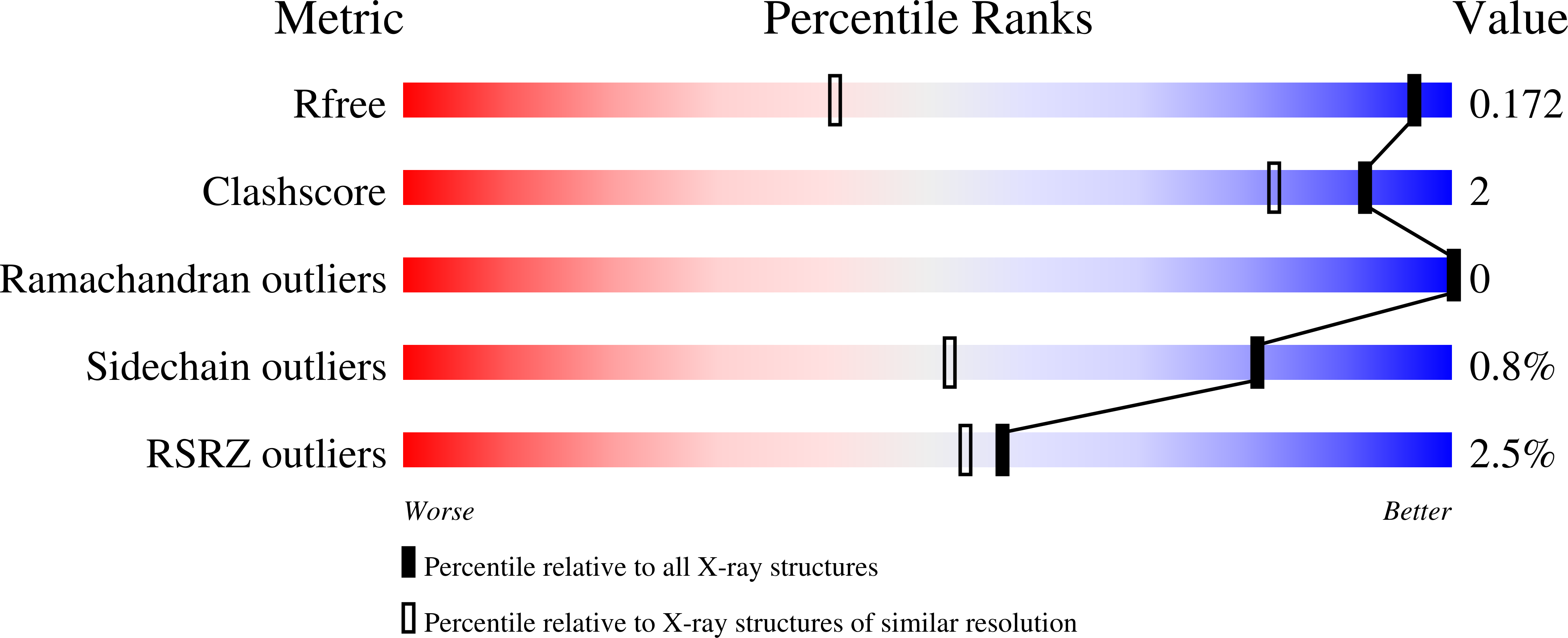
Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
Nuti, E., Cuffaro, D., Bernardini, E., Camodeca, C., Panelli, L., Chaves, S., Ciccone, L., Tepshi, L., Vera, L., Orlandini, E., Nencetti, S., Stura, E.A., Santos, M.A., Dive, V., Rossello, A.(2018) J Med Chem 61: 4421-4435
- PubMed: 29727184
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00096
- Primary Citation of Related Structures:
6EKN, 6ELA, 6ENM, 6EOX, 6ESM - PubMed Abstract:
Matrix metalloproteinase-12 (MMP-12) selective inhibitors could play a role in the treatment of lung inflammatory and cardiovascular diseases. In the present study, the previously reported 4-methoxybiphenylsulfonyl hydroxamate and carboxylate based inhibitors (1b and 2b) were modified to enhance their selectivity for MMP-12. In the newly synthesized thioaryl derivatives, the nature of the zinc binding group (ZBG) and the sulfur oxidation state were changed. Biological assays carried out in vitro on human MMPs with the resulting compounds led to identification of a sulfide, 4a, bearing an N-1-hydroxypiperidine-2,6-dione (HPD) group as new ZBG. Compound 4a is a promising hit compound since it displayed a nanomolar affinity for MMP-12 with a marked selectivity over MMP-9, MMP-1, and MMP-14. Solution complexation studies with Zn 2+ were performed to characterize the chelating abilities of the new compounds and confirmed the bidentate binding mode of HPD derivatives. X-ray crystallography studies using MMP-12 and MMP-9 catalytic domains were carried out to rationalize the biological results.
Organizational Affiliation:
Dipartimento di Farmacia , Università di Pisa , Via Bonanno 6 , 56126 Pisa , Italy.