Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
Macias, A.T., Williamson, D.S., Allen, N., Borgognoni, J., Clay, A., Daniels, Z., Dokurno, P., Drysdale, M.J., Francis, G.L., Graham, C.J., Howes, R., Matassova, N., Murray, J.B., Parsons, R., Shaw, T., Surgenor, A.E., Terry, L., Wang, Y., Wood, M., Massey, A.J.(2011) J Med Chem 54: 4034-4041
- PubMed: 21526763
- DOI: https://doi.org/10.1021/jm101625x
- Primary Citation of Related Structures:
3LDL, 3LDN, 3LDO, 3LDP, 3LDQ, 3M3Z - PubMed Abstract:
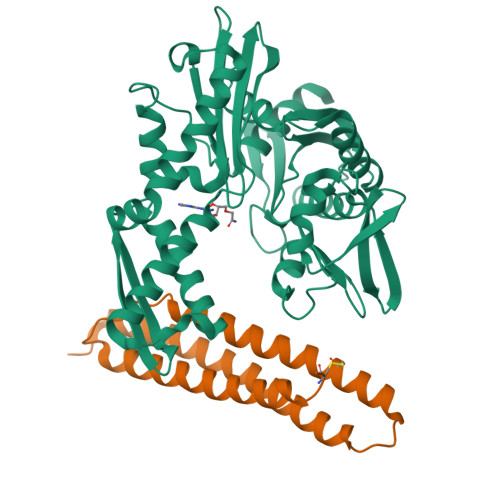
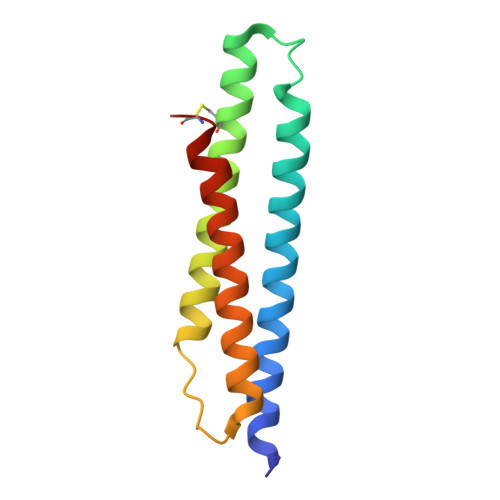
78 kDa glucose-regulated protein (Grp78) is a heat shock protein (HSP) involved in protein folding that plays a role in cancer cell proliferation. Binding of adenosine-derived inhibitors to Grp78 was characterized by surface plasmon resonance and isothermal titration calorimetry. The most potent compounds were 13 (VER-155008) with K(D) = 80 nM and 14 with K(D) = 60 nM. X-ray crystal structures of Grp78 bound to ATP, ADPnP, and adenosine derivative 10 revealed differences in the binding site between Grp78 and homologous proteins.
Organizational Affiliation:
Vernalis (R&D) Ltd., Granta Park, Great Abington, Cambridge, CB21 6GB, UK. a.macias@vernalis.com