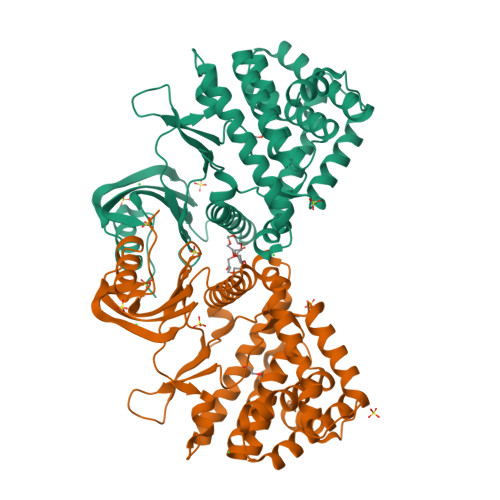
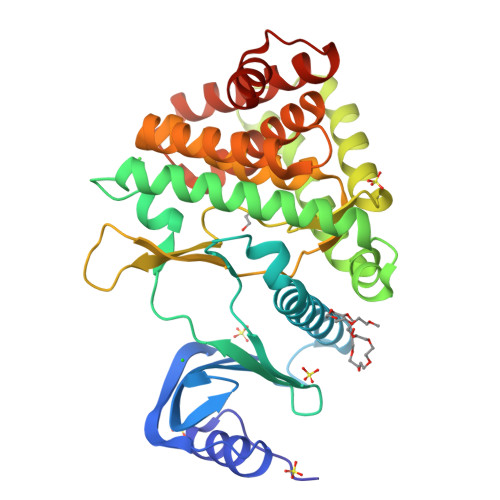
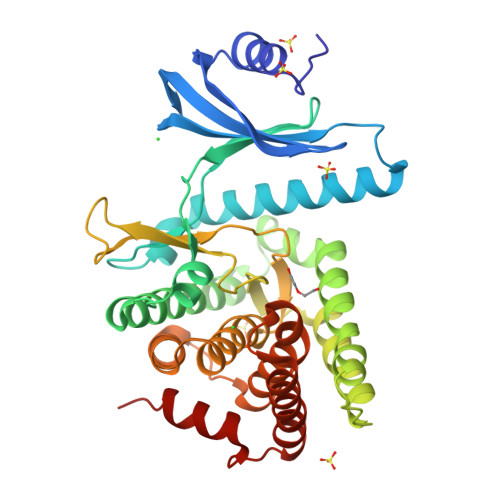
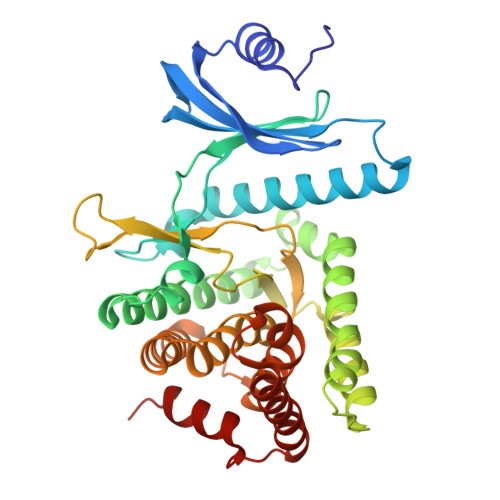
Substrate recognition by the 4-hydroxytryptamine kinase PsiK in psilocybin biosynthesis.
Rogge, K., Wagner, T.J., Hoffmeister, D., Rupp, B., Werten, S.(2025) FEBS Lett 599: 447-455
- PubMed: 39449146
- DOI: https://doi.org/10.1002/1873-3468.15042
- Primary Citation of Related Structures:
9ETO - PubMed Abstract:
Psilocybin, the natural hallucinogen from Psilocybe (magic) mushrooms, is a highly promising drug candidate for the treatment of depression and several other mental health conditions. Biosynthesis of psilocybin from the amino acid l-tryptophan involves four strictly sequential modifications. The third of these, ATP-dependent phosphorylation of the intermediate 4-hydroxytryptamine, is catalysed by PsiK. Here we present a crystallographic analysis and a structure-based mutagenesis study of this kinase, providing insight into its mode of substrate recognition. The results of our work will support future bioengineering efforts aimed at generating variants of psilocybin with enhanced therapeutic properties.
Organizational Affiliation:
Institute of Pharmacy, Friedrich Schiller University, Jena, Germany.