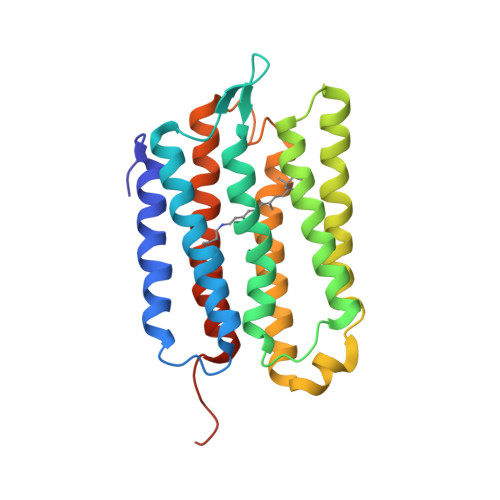
Membrane protein structure determination from Paramagnetic Relaxation Enhancement and internuclear distance restraints.
Vaz, R.F., Brown, L.S., Ladizhansky, V.(2025) J Biomol NMR
- PubMed: 40156665
- DOI: https://doi.org/10.1007/s10858-025-00467-w
- Primary Citation of Related Structures:
9MEM - PubMed Abstract:
Magic angle spinning nuclear magnetic resonance (MAS NMR) is well suited for the determination of protein structure. The key structural information is obtained in the form of spectral cross peaks between spatially close nuclear spins, but assigning these cross peaks unambiguously to unique spin pairs is often a tedious task because of spectral overlap. Here, we use a seven-helical membrane protein Anabaena Sensory Rhodopsin (ASR) as a model system to demonstrate that transverse Paramagnetic Relaxation Enhancements (PRE) extracted from 2D MAS NMR spectra could be used to obtain a protein structural model. Starting with near complete assignments (93%) of ASR residues, TALOS + predicted backbone dihedral angles and secondary structure restraints in the form of backbone hydrogen bonds are combined with PRE-based restraints and used to generate a coarse model. This model is subsequently utilized as a template reference to facilitate automated assignments of highly ambiguous internuclear correlations. The template is used in an iterative cross peak assignment process and is progressively improved through the inclusion of disambiguated restraints, thereby converging to a low root-mean-square-deviation structural model. In addition to improving structure calculation conversion, the inclusion of PREs also improves packing between helices within an alpha-helical bundle.
Organizational Affiliation:
Department of Physics and Biophysics Interdepartmental Group, University of Guelph, 50 Stone Rd. E., Guelph, ON, N1G 2W1, Canada.