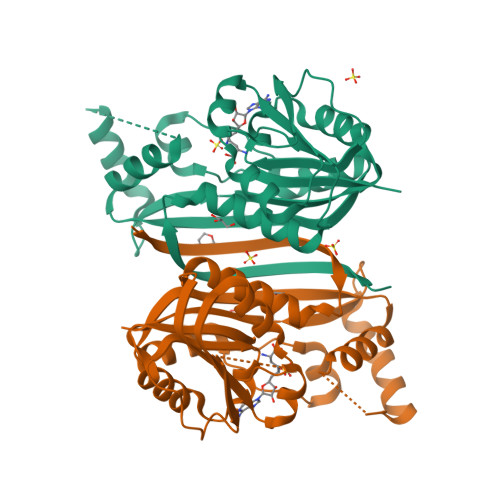
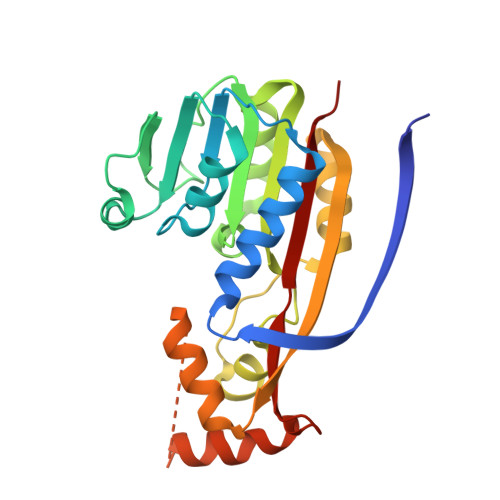
A bacterial methyltransferase that initiates biotin synthesis, an attractive anti-ESKAPE druggable pathway.
Su, Z., Zhang, W., Shi, Y., Cui, T., Xu, Y., Yang, R., Huang, M., Zhou, C., Zhang, H., Lu, T., Qu, J., He, Z.G., Gan, J., Feng, Y.(2024) Sci Adv 10: eadp3954-eadp3954
- PubMed: 39705367
- DOI: https://doi.org/10.1126/sciadv.adp3954
- Primary Citation of Related Structures:
8X8I, 8X8J - PubMed Abstract:
The covalently attached cofactor biotin plays pivotal roles in central metabolism. The top-priority ESKAPE-type pathogens, Acinetobacter baumannii and Klebsiella pneumoniae , constitute a public health challenge of global concern. Despite the fact that the late step of biotin synthesis is a validated anti-ESKAPE drug target, the primary stage remains fragmentarily understood. We report the functional definition of two BioC isoenzymes (AbBioC for A. baumannii and KpBioC for K. pneumoniae ) that act as malonyl-ACP methyltransferase and initiate biotin synthesis. The physiological requirement of biotin is diverse within ESKAPE pathogens. CRISPR-Cas9-based inactivation of bioC rendered A. baumannii and K. pneumoniae biotin auxotrophic. The availability of soluble AbBioC enabled the in vitro reconstitution of DTB/biotin synthesis. We solved two crystal structures of AbBioC bound to SAM cofactor (2.54 angstroms) and sinefungin (SIN) inhibitor (1.72 angstroms). Structural and functional study provided molecular basis for SIN inhibition of BioC. We demonstrated that BioC methyltransferase plays dual roles in K. pneumoniae infection and A. baumannii colistin resistance.
Organizational Affiliation:
Key Laboratory of Multiple Organ Failure (Ministry of Education), and Departments of Microbiology and General Intensive Care Unit of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310058, China.