Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Minen, R.I., Bhayani, J.A., Hartman, M.D., Cereijo, A.E., Zheng, Y., Ballicora, M.A., Iglesias, A.A., Liu, D., Figueroa, C.M.(2022) Plant Cell Physiol 63: 658-670
- PubMed: 35243499
- DOI: https://doi.org/10.1093/pcp/pcac029
- Primary Citation of Related Structures:
7S5F, 7S5I - PubMed Abstract:
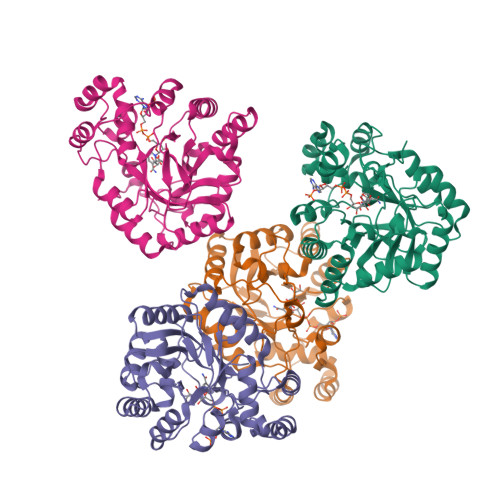
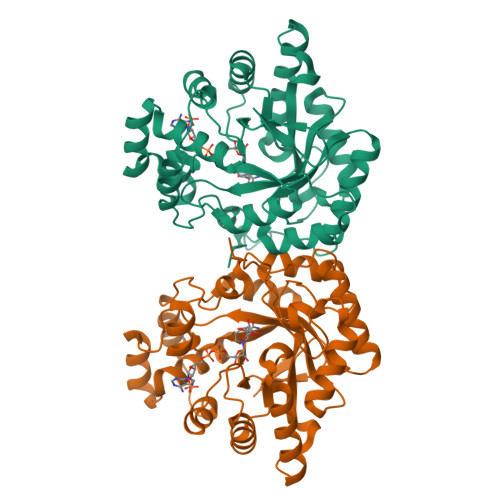
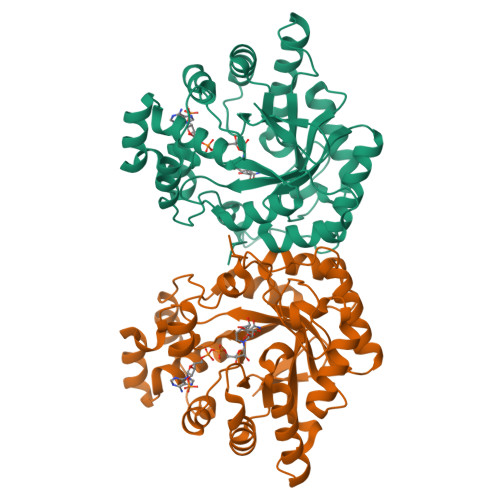
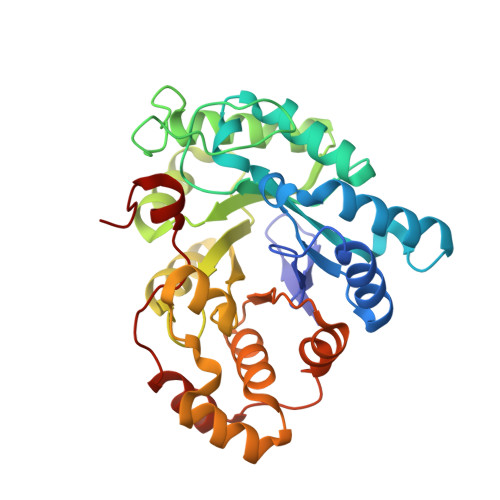
Sugar alcohols are major photosynthetic products in plant species from the Apiaceae and Plantaginaceae families. Mannose-6-phosphate reductase (Man6PRase) and aldose-6-phosphate reductase (Ald6PRase) are key enzymes for synthesizing mannitol and glucitol in celery (Apium graveolens) and peach (Prunus persica), respectively. In this work, we report the first crystal structures of dimeric plant aldo/keto reductases (AKRs), celery Man6PRase (solved in the presence of mannonic acid and NADP+) and peach Ald6PRase (obtained in the apo form). Both structures displayed the typical TIM barrel folding commonly observed in proteins from the AKR superfamily. Analysis of the Man6PRase holo form showed that residues putatively involved in the catalytic mechanism are located close to the nicotinamide ring of NADP+, where the hydride transfer to the sugar phosphate should take place. Additionally, we found that Lys48 is important for the binding of the sugar phosphate. Interestingly, the Man6PRase K48A mutant had a lower catalytic efficiency with mannose-6-phosphate but a higher catalytic efficiency with mannose than the wild type. Overall, our work sheds light on the structure-function relationships of important enzymes to synthesize sugar alcohols in plants.
Organizational Affiliation:
UNL, CONICET, FBCB, Instituto de Agrobiotecnología del Litoral, UNL, CONICET, FBCB, Santa Fe 3000, Argentina.