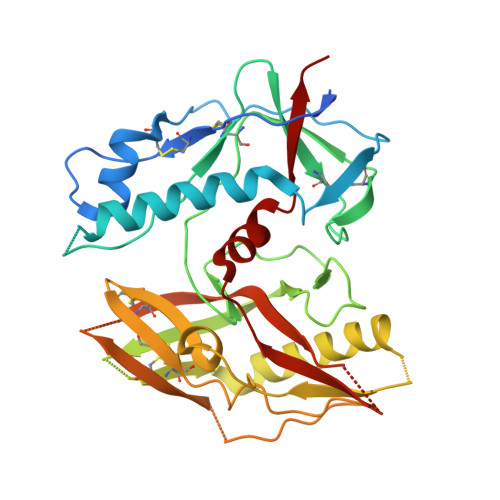
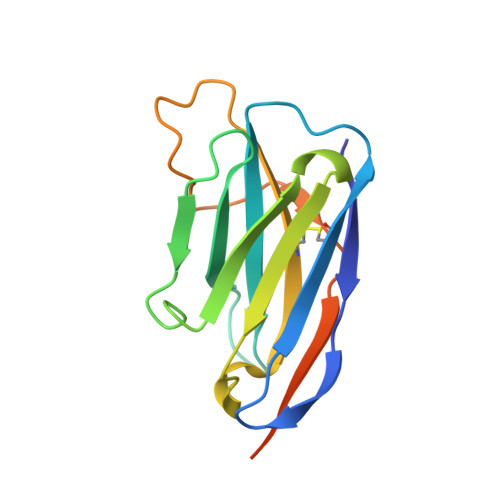
Structural basis for llama nanobody recognition and neutralization of HIV-1 at the CD4-binding site.
Zhou, T., Chen, L., Gorman, J., Wang, S., Kwon, Y.D., Lin, B.C., Louder, M.K., Rawi, R., Stancofski, E.D., Yang, Y., Zhang, B., Quigley, A.F., McCoy, L.E., Rutten, L., Verrips, T., Weiss, R.A., Doria-Rose, N.A., Shapiro, L., Kwong, P.D.(2022) Structure 30: 862-875.e4
- PubMed: 35413243
- DOI: https://doi.org/10.1016/j.str.2022.03.012
- Primary Citation of Related Structures:
7LPN, 7R73, 7R74, 7RI1, 7RI2 - PubMed Abstract:
Nanobodies can achieve remarkable neutralization of genetically diverse pathogens, including HIV-1. To gain insight into their recognition, we determined crystal structures of four llama nanobodies (J3, A12, C8, and D7), all of which targeted the CD4-binding site, in complex with the HIV-1 envelope (Env) gp120 core, and determined a cryoelectron microscopy (cryo-EM) structure of J3 with the Env trimer. Crystal and cryo-EM structures of J3 complexes revealed this nanobody to mimic binding to the prefusion-closed trimer for the primary site of CD4 recognition as well as a secondary quaternary site. In contrast, crystal structures of A12, C8, and D7 with gp120 revealed epitopes that included portions of the gp120 inner domain, inaccessible on the prefusion-closed trimer. Overall, these structures explain the broad and potent neutralization of J3 and limited neutralization of A12, C8, and D7, which utilized binding modes incompatible with the neutralization-targeted prefusion-closed conformation of Env.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.