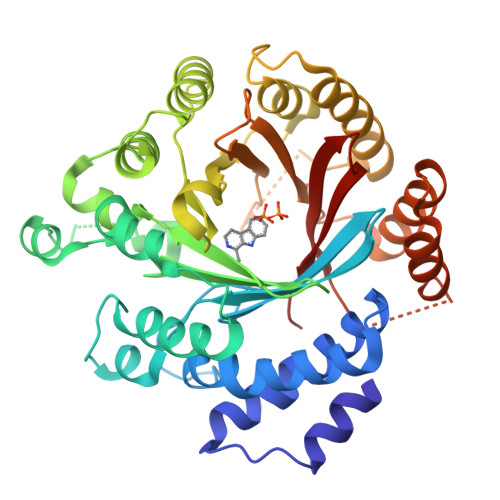
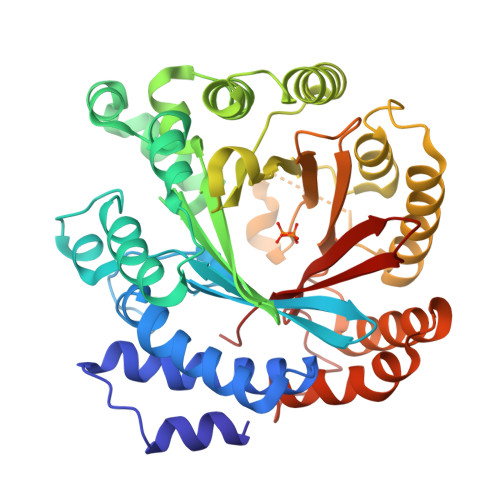
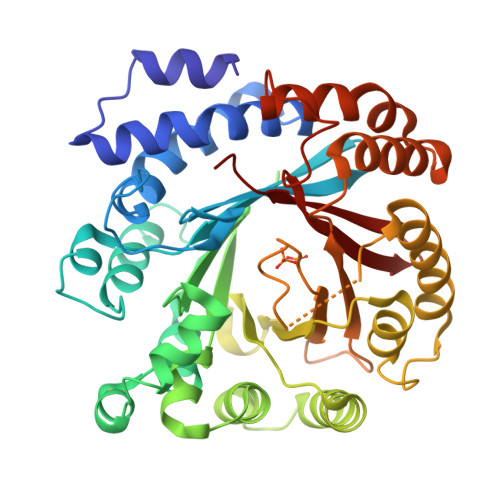
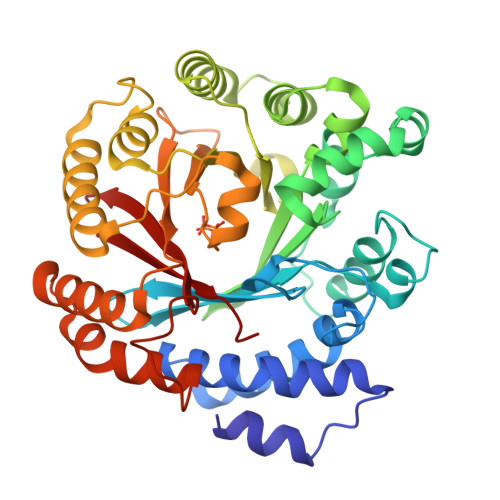
Catalytic potential of a fungal indole prenyltransferase toward beta-carbolines, harmine and harman, and their prenylation effects on antibacterial activity.
Hamdy, S.A., Kodama, T., Nakashima, Y., Han, X., Morita, H.(2022) J Biosci Bioeng 134: 311-317
- PubMed: 35931602
- DOI: https://doi.org/10.1016/j.jbiosc.2022.07.004
- Primary Citation of Related Structures:
7Y3V - PubMed Abstract:
The prenylation of compounds has attracted much attention, since it often adds bioactivity to non-prenylated compounds. We employed an enzyme assay with CdpNPT, an indole prenyltransferase from Aspergillus fumigatus with two naturally occurring β-carbolines, harmine (3) and harman (4) as prenyl acceptors, in the presence of dimethylallyl diphosphate (DMAPP) as the prenyl donor. The enzyme accepted these two prenyl acceptor substrates to produce 6-(3',3'-dimethylallyl)harmine (5) from 3 and 9-(3',3'-dimethylallyl)harman (6) and 6-(3',3'-dimethylallyl)harman (7) from 4. The X-ray crystal structure analysis of the CdpNPT (38-440) truncated mutant complexed with 4, and docking simulation studies of DMAPP to the crystal structure of the CdpNPT (38-440) mutant, suggested that CdpNPT could employ the two-step prenylation mechanism to produce 7, while the enzyme produced 6 with either one- or two-step prenylation mechanisms. Furthermore, the antibacterial assays revealed that the 3',3'-dimethylallylation of 3 and 4, as well as harmol (1), at C-6 enhanced the activities against Staphylococcus aureus and Bacillus subtilis.
Organizational Affiliation:
Department of Pharmacognosy, Faculty of Pharmacy, Cairo University, Kasr El-Aini St., Cairo 11562, Egypt; Institute of Natural Medicine, University of Toyama, 2630 Sugitani, Toyama 930-0194, Japan.