Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Raja Reddy, K., Totrov, M., Lomovskaya, O., Griffith, D.C., Tarazi, Z., Clifton, M.C., Hecker, S.J.(2022) Bioorg Med Chem 62: 116722-116722
- PubMed: 35358864
- DOI: https://doi.org/10.1016/j.bmc.2022.116722
- Primary Citation of Related Structures:
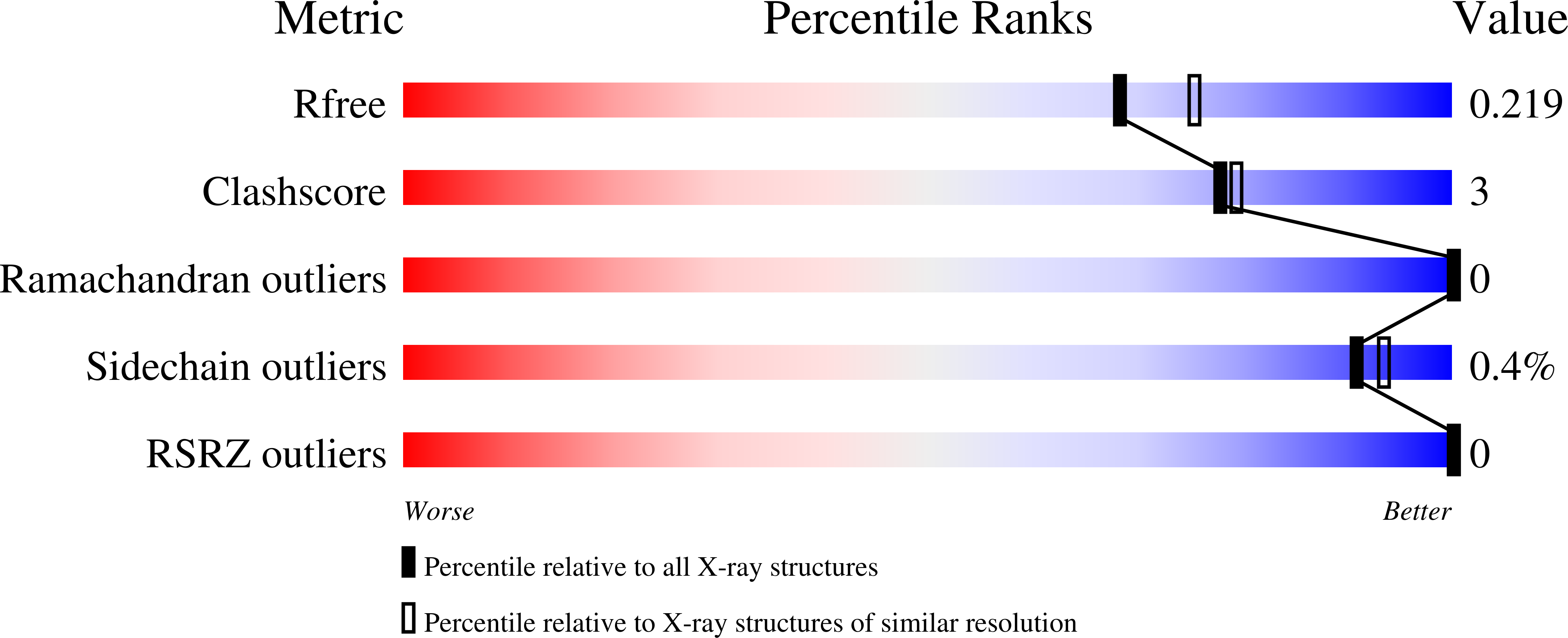
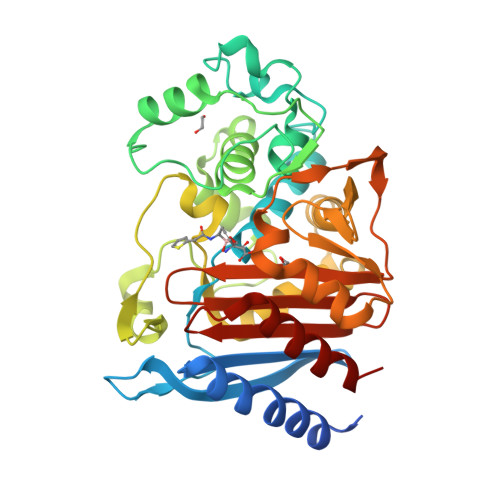
7TI0, 7TI1, 7TI2 - PubMed Abstract:
Early efforts to broaden the spectrum and potency of cyclic boronic acid β-lactamase inhibitor vaborbactam included a series of 7-membered ring boronates. Exploration of stereoisomers and incorporation of heteroatoms allowed identification of the all-carbon cyclic boronate with substituents trans as the preferred core structure, showing inhibition of Class A and C enzymes. Crystal structures of one analog bound to important β-lactamase enzymes were obtained. When isolated under acidic conditions, these compounds spontaneously formed a neutral cyclic anhydride (intramolecular prodrug) which was shown to have much-improved oral bioavailability (52-69%) compared to the ring-opened carboxylate salt (9%).
Organizational Affiliation:
Qpex Biopharma, Inc, 6275 Nancy Ridge Dr., Suite 100, San Diego, CA 92121, United States.