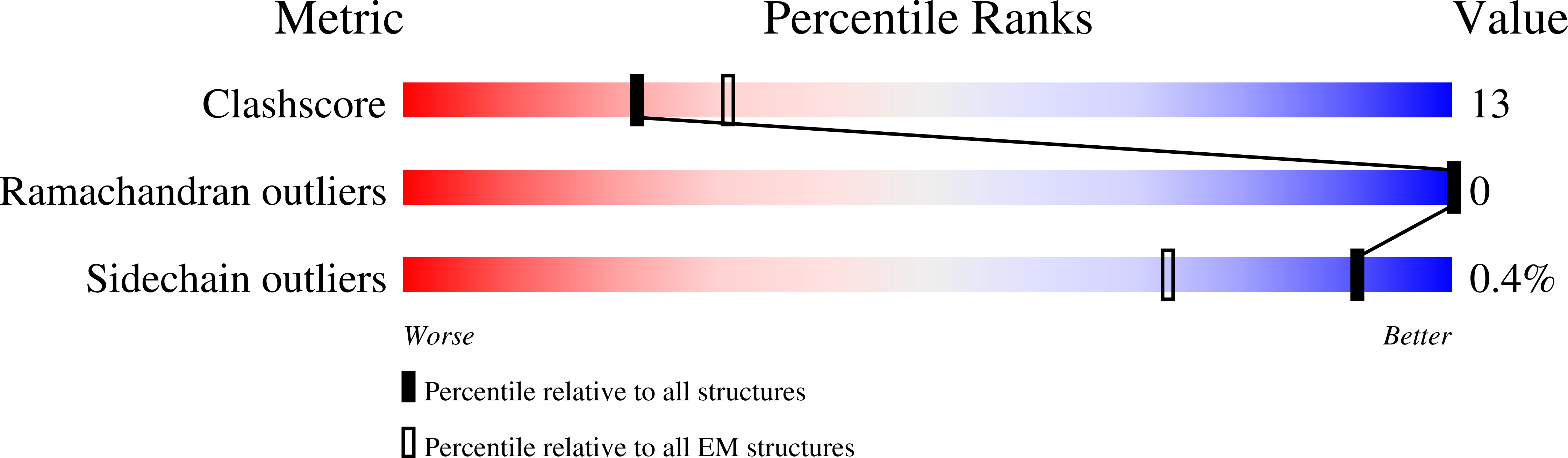Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation.
Klink, B.U., Herrmann, E., Antoni, C., Langemeyer, L., Kiontke, S., Gatsogiannis, C., Ungermann, C., Raunser, S., Kummel, D.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35105815
- DOI: https://doi.org/10.1073/pnas.2121494119
- Primary Citation of Related Structures:
7QLA - PubMed Abstract:
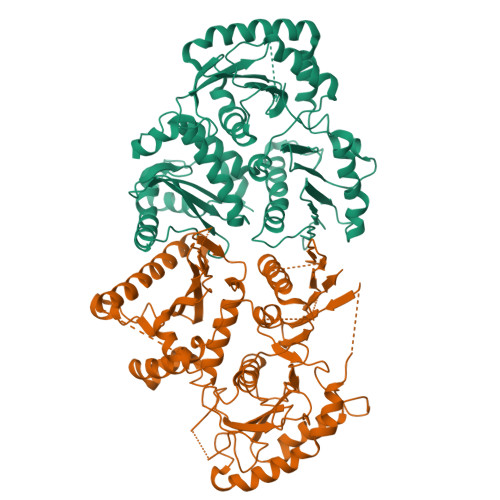
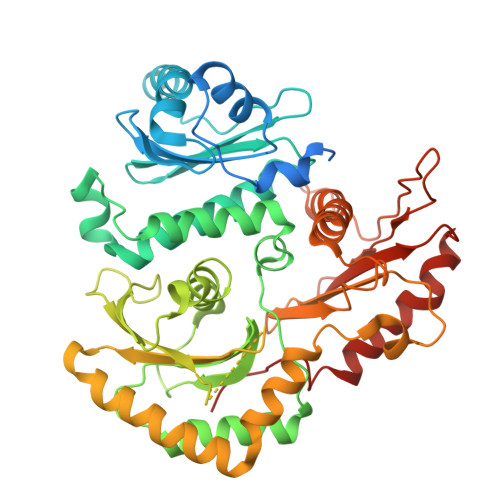
Activation of the GTPase Rab7/Ypt7 by its cognate guanine nucleotide exchange factor (GEF) Mon1-Ccz1 marks organelles such as endosomes and autophagosomes for fusion with lysosomes/vacuoles and degradation of their content. Here, we present a high-resolution cryogenic electron microscopy structure of the Mon1-Ccz1 complex that reveals its architecture in atomic detail. Mon1 and Ccz1 are arranged side by side in a pseudo-twofold symmetrical heterodimer. The three Longin domains of each Mon1 and Ccz1 are triangularly arranged, providing a strong scaffold for the catalytic center of the GEF. At the opposite side of the Ypt7-binding site, a positively charged and relatively flat patch stretches the Longin domains 2/3 of Mon1 and functions as a phosphatidylinositol phosphate-binding site, explaining how the GEF is targeted to membranes. Our work provides molecular insight into the mechanisms of endosomal Rab activation and serves as a blueprint for understanding the function of members of the Tri Longin domain Rab-GEF family.
Organizational Affiliation:
Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, 44227 Dortmund, Germany.