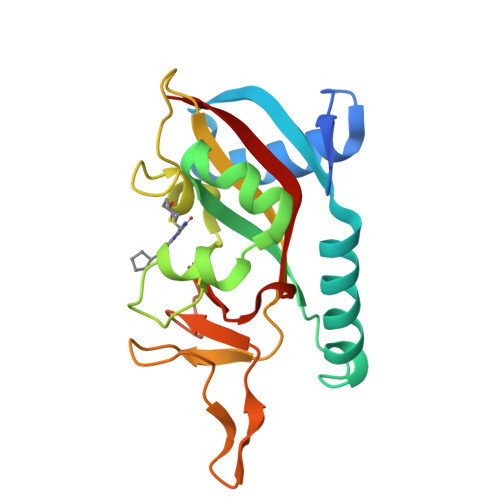
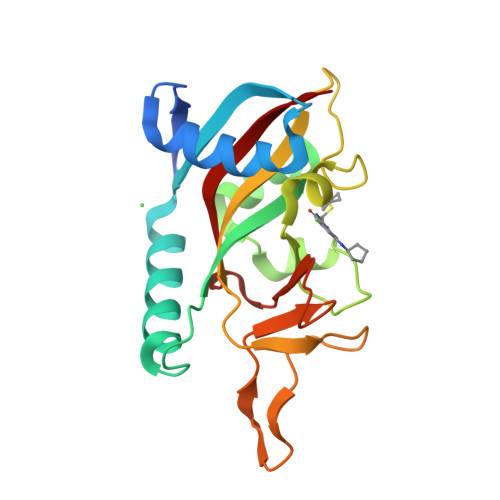
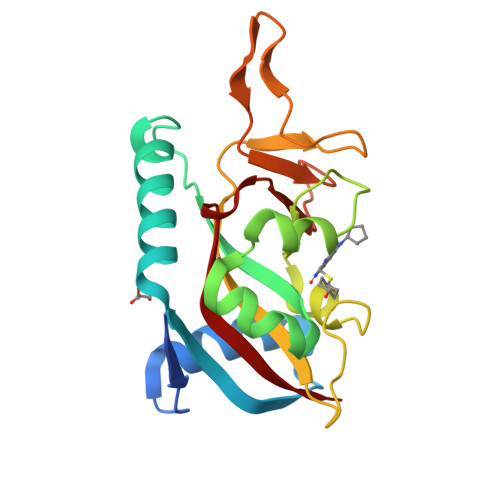
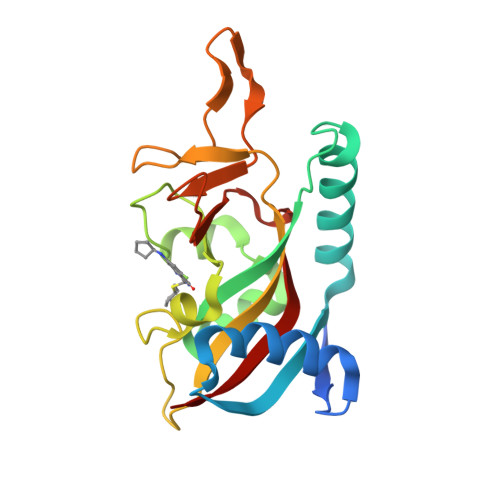
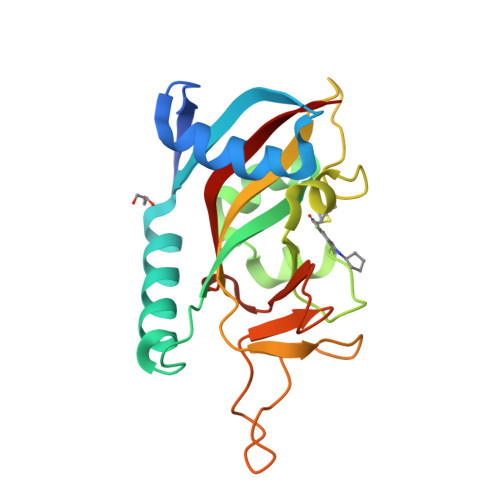
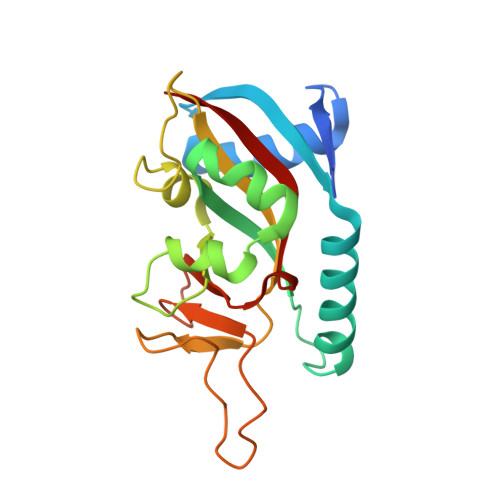
Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Wigle, T.J., Ren, Y., Molina, J.R., Blackwell, D.J., Schenkel, L.B., Swinger, K.K., Kuplast-Barr, K., Majer, C.R., Church, W.D., Lu, A.Z., Mo, J., Abo, R., Cheung, A., Dorsey, B.W., Niepel, M., Perl, N.R., Vasbinder, M.M., Keilhack, H., Kuntz, K.W.(2021) Chembiochem 22: 2107-2110
- PubMed: 33838082
- DOI: https://doi.org/10.1002/cbic.202100047
- Primary Citation of Related Structures:
7L9Y, 7LUN - PubMed Abstract:
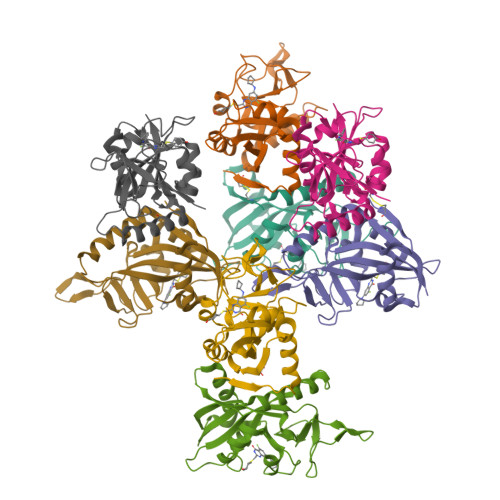
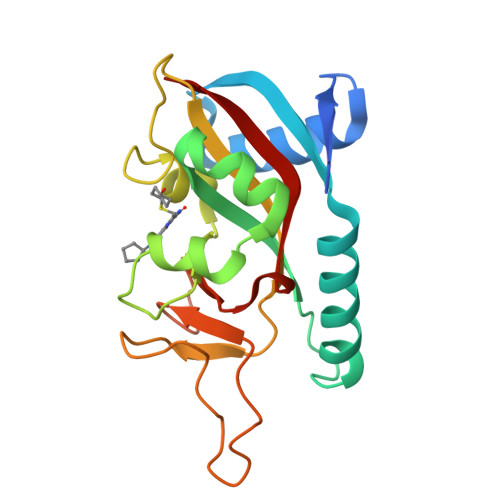
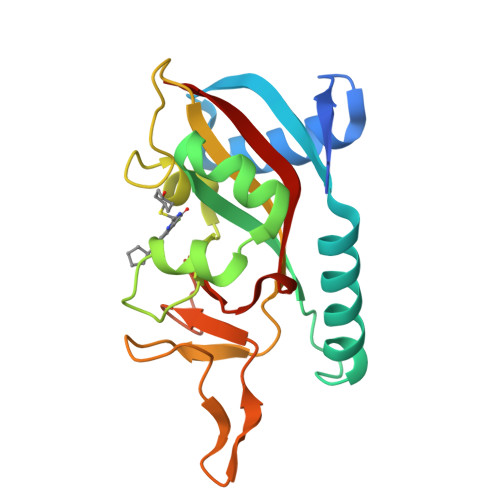
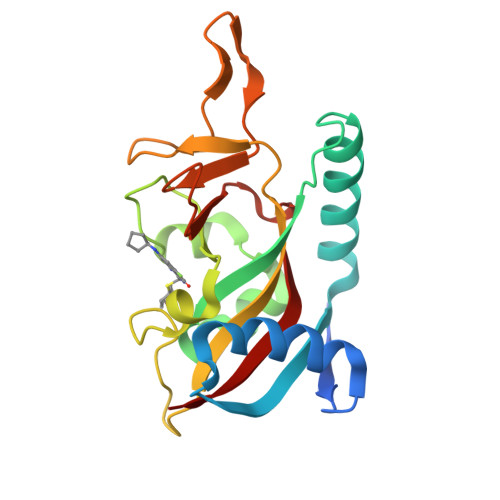
PARP14 is an interferon-stimulated gene that is overexpressed in multiple tumor types, influencing pro-tumor macrophage polarization as well as suppressing the antitumor inflammation response by modulating IFN-γ and IL-4 signaling. PARP14 is a 203 kDa protein that possesses a catalytic domain responsible for the transfer of mono-ADP-ribose to its substrates. PARP14 also contains three macrodomains and a WWE domain which are binding modules for mono-ADP-ribose and poly-ADP-ribose, respectively, in addition to two RNA recognition motifs. Catalytic inhibitors of PARP14 have been shown to reverse IL-4 driven pro-tumor gene expression in macrophages, however it is not clear what roles the non-enzymatic biomolecular recognition motifs play in PARP14-driven immunology and inflammation. To further understand this, we have discovered a heterobifunctional small molecule designed based on a catalytic inhibitor of PARP14 that binds in the enzyme's NAD + -binding site and recruits cereblon to ubiquitinate it and selectively target it for degradation.
Organizational Affiliation:
Ribon Therapeutics, 35 Cambridgepark Dr., Suite 300, Cambridge, MA 02140, USA.