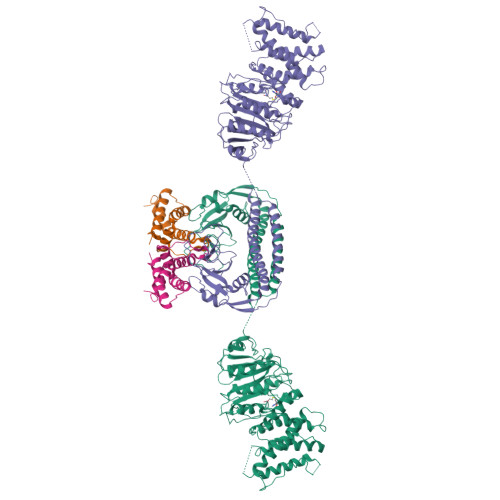
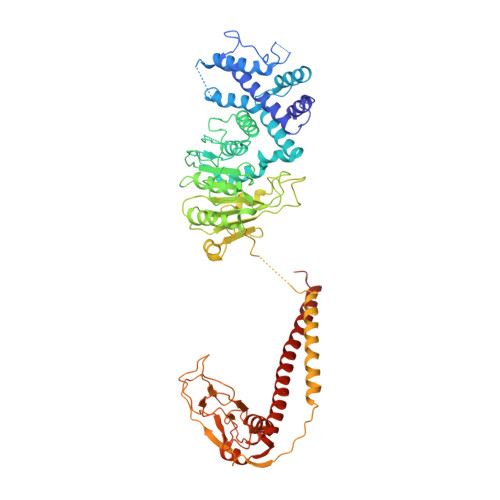
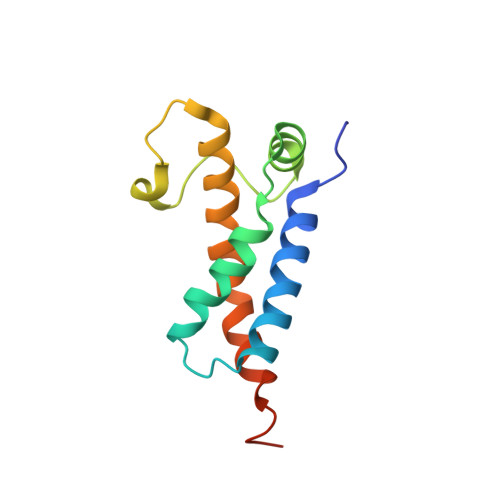
Structural features of a minimal intact methyltransferase of a type I restriction-modification system.
Seo, P.W., Hofmann, A., Kim, J.H., Hwangbo, S.A., Kim, J.H., Kim, J.W., Huynh, T.Y.L., Choy, H.E., Kim, S.J., Lee, J., Lee, J.O., Jin, K.S., Park, S.Y., Kim, J.S.(2022) Int J Biol Macromol 208: 381-389
- PubMed: 35337914
- DOI: https://doi.org/10.1016/j.ijbiomac.2022.03.115
- Primary Citation of Related Structures:
7EEW - PubMed Abstract:
Type I restriction-modification enzymes are oligomeric proteins composed of methylation (M), DNA sequence-recognition (S), and restriction (R) subunits. The different bipartite DNA sequences of 2-4 consecutive bases are recognized by two discerned target recognition domains (TRDs) located at the two-helix bundle of the two conserved regions (CRs). Two M-subunits and a single S-subunit form an oligomeric protein that functions as a methyltransferase (M 2 S 1 MTase). Here, we present the crystal structure of the intact MTase from Vibrio vulnificus YJ016 in complex with the DNA-mimicking Ocr protein and the S-adenosyl-L-homocysteine (SAH). This MTase includes the M-domain with a helix tail (M-tail helix) and the S 1/2 -domain of a TRD and a CR α-helix. The Ocr binds to the cleft of the TRD surface and SAH is located in the pocket within the M-domain. The solution- and negative-staining electron microscopy-based reconstructed (M 1 S 1/2 ) 2 structure reveals a symmetric (S 1/2 ) 2 assembly using two CR-helices and two M-tail helices as a pivot, which is plausible for recognizing two DNA regions of same sequence. The conformational flexibility of the minimal M 1 S 1/2 MTase dimer indicates a particular state resembling the structure of M 2 S 1 MTases.
Organizational Affiliation:
Department of Chemistry, Chonnam National University, Gwangju 61186, Republic of Korea.