Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Chiniadis, L., Giastas, P., Bratsos, I., Papakyriakou, A.(2021) Inorg Chem 60: 10729-10737
- PubMed: 34197115
- DOI: https://doi.org/10.1021/acs.inorgchem.1c01441
- Primary Citation of Related Structures:
7BCU, 7BCX, 7BD0, 7BDM - PubMed Abstract:
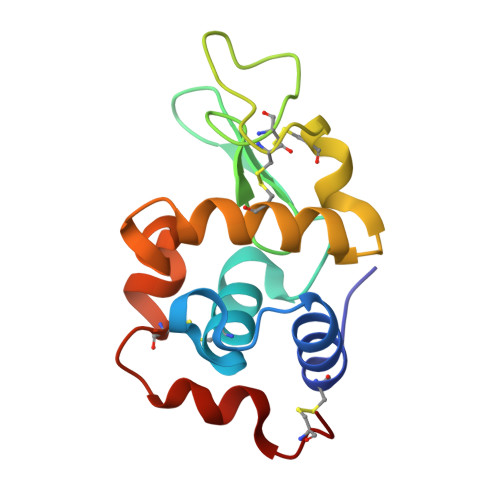
The pharmacological profile of medicinally relevant Ru(III) coordination compounds has been ascribed to their interactions with proteins, as several studies have provided evidence that DNA is not the primary target. In this regard, numerous spectroscopic and crystallographic studies have indicated that the Ru(III) ligands play an important role in determining the metal binding site, acting as the recognition element in the early stages of the protein-complex formation. Herein, we present a series of near-atomic-resolution X-ray crystal structures of the adducts formed between the antimetastatic metallodrug imidazolium trans -[tetrachlorido( S -dimethyl sufoxide)(1 H -imidazole)ruthenate(III)] ( NAMI-A ) and hen egg-white lysozyme (HEWL). These structures elucidate a series of binding events starting from the noncovalent interaction of intact NAMI-A ions with HEWL (1.5 h), followed by the stepwise exchange of all Ru ligands except for 1 H -imidazole (26 h) to the final "ruthenated" protein comprising one aquated Ru ion coordinated to histidine-15 of HEWL (98 h). Our structural data clearly support a two-step mechanism of protein ruthenation, illustrating the ligand-mediated recognition step of the process.
Organizational Affiliation:
Department of Neurobiology, Hellenic Pasteur Institute, 11521 Athens, Greece.