Peptide Inhibitors of the alpha-Cobratoxin-Nicotinic Acetylcholine Receptor Interaction.
Lynagh, T., Kiontke, S., Meyhoff-Madsen, M., Gless, B.H., Johannesen, J., Kattelmann, S., Christiansen, A., Dufva, M., Laustsen, A.H., Devkota, K., Olsen, C.A., Kummel, D., Pless, S.A., Lohse, B.(2020) J Med Chem 63: 13709-13718
- PubMed: 33143415
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01202
- Primary Citation of Related Structures:
6ZFM - PubMed Abstract:
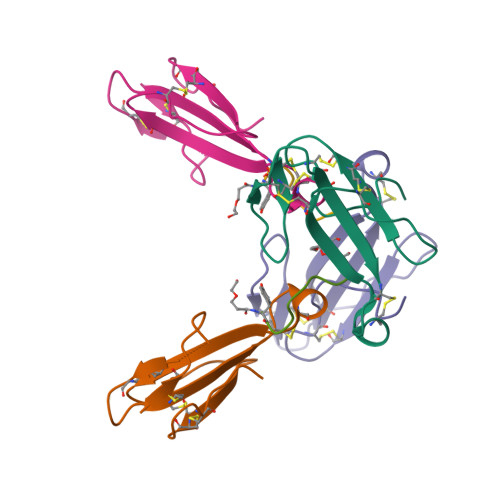
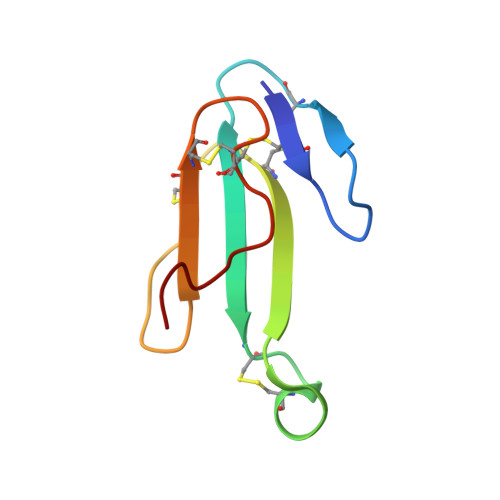
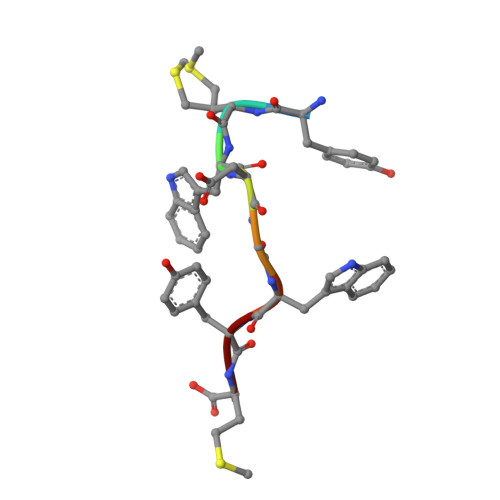
Venomous snakebites cause >100 000 deaths every year, in many cases via potent depression of human neuromuscular signaling by snake α-neurotoxins. Emergency therapy still relies on antibody-based antivenom, hampered by poor access, frequent adverse reactions, and cumbersome production/purification. Combining high-throughput discovery and subsequent structure-function characterization, we present simple peptides that bind α-cobratoxin (α-Cbtx) and prevent its inhibition of nicotinic acetylcholine receptors (nAChRs) as a lead for the development of alternative antivenoms. Candidate peptides were identified by phage display and deep sequencing, and hits were characterized by electrophysiological recordings, leading to an 8-mer peptide that prevented α-Cbtx inhibition of nAChRs. We also solved the peptide:α-Cbtx cocrystal structure, revealing that the peptide, although of unique primary sequence, binds to α-Cbtx by mimicking structural features of the nAChR binding pocket. This demonstrates the potential of small peptides to neutralize lethal snake toxins in vitro, establishing a potential route to simple, synthetic, low-cost antivenoms.
Organizational Affiliation:
Sars International Centre for Marine Molecular Biology, University of Bergen, Thormøhlensgate 55, 5008 Bergen, Norway.