Identification ofN-Methyl Nicotinamide andN-Methyl Pyridazine-3-Carboxamide Pseudokinase Domain Ligands as Highly Selective Allosteric Inhibitors of Tyrosine Kinase 2 (TYK2).
Moslin, R., Zhang, Y., Wrobleski, S.T., Lin, S., Mertzman, M., Spergel, S., Tokarski, J.S., Strnad, J., Gillooly, K., McIntyre, K.W., Zupa-Fernandez, A., Cheng, L., Sun, H., Chaudhry, C., Huang, C., D'Arienzo, C., Heimrich, E., Yang, X., Muckelbauer, J.K., Chang, C., Tredup, J., Mulligan, D., Xie, D., Aranibar, N., Chiney, M., Burke, J.R., Lombardo, L., Carter, P.H., Weinstein, D.S.(2019) J Med Chem 62: 8953-8972
- PubMed: 31314518
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00443
- Primary Citation of Related Structures:
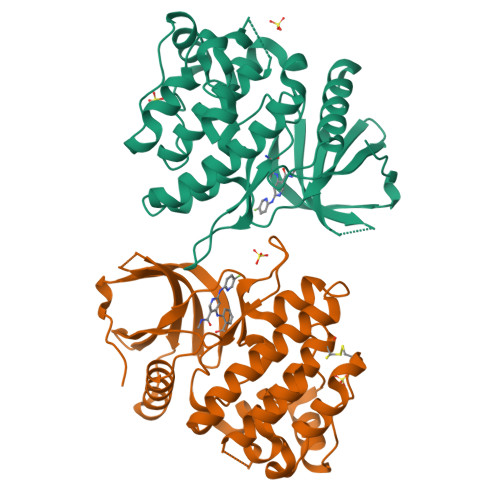
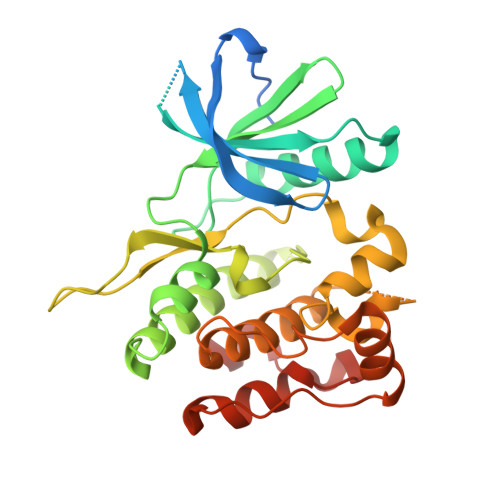
6NZE, 6NZF, 6NZH - PubMed Abstract:
As a member of the Janus (JAK) family of nonreceptor tyrosine kinases, TYK2 plays an important role in mediating the signaling of pro-inflammatory cytokines including IL-12, IL-23, and type 1 interferons. The nicotinamide 4 , identified by a SPA-based high-throughput screen targeting the TYK2 pseudokinase domain, potently inhibits IL-23 and IFNα signaling in cellular assays. The described work details the optimization of this poorly selective hit ( 4 ) to potent and selective molecules such as 47 and 48 . The discoveries described herein were critical to the eventual identification of the clinical TYK2 JH2 inhibitor (see following report in this issue). Compound 48 provided robust inhibition in a mouse IL-12-induced IFNγ pharmacodynamic model as well as efficacy in an IL-23 and IL-12-dependent mouse colitis model. These results demonstrate the ability of TYK2 JH2 domain binders to provide a highly selective alternative to conventional TYK2 orthosteric inhibitors.