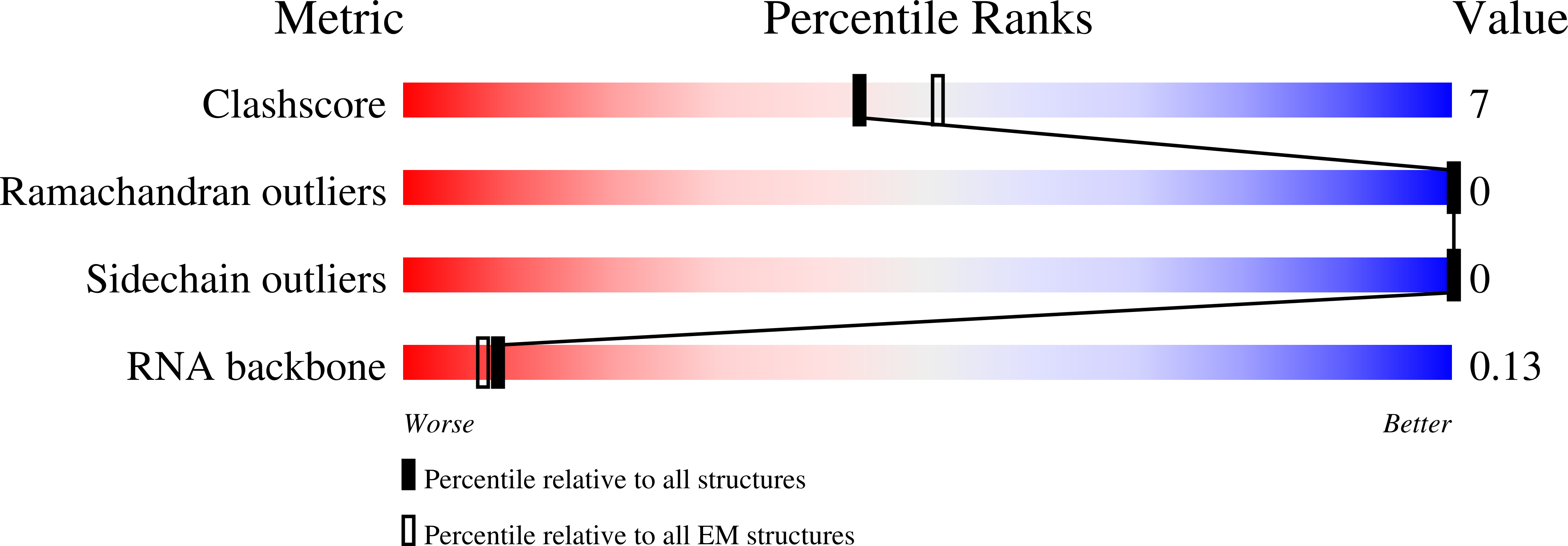
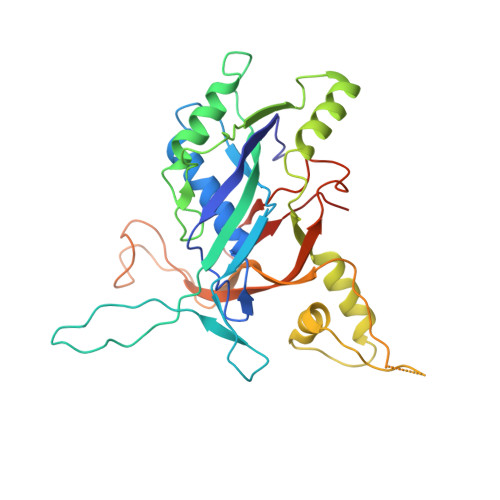
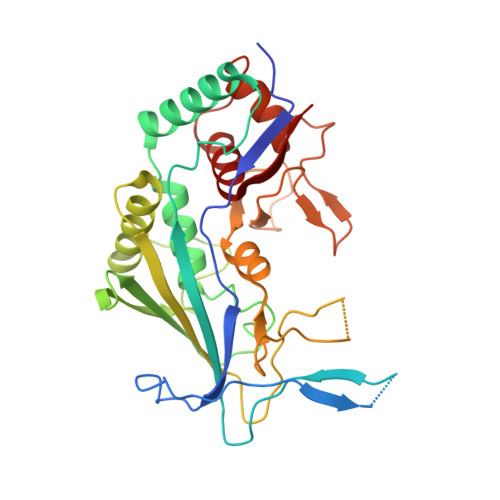
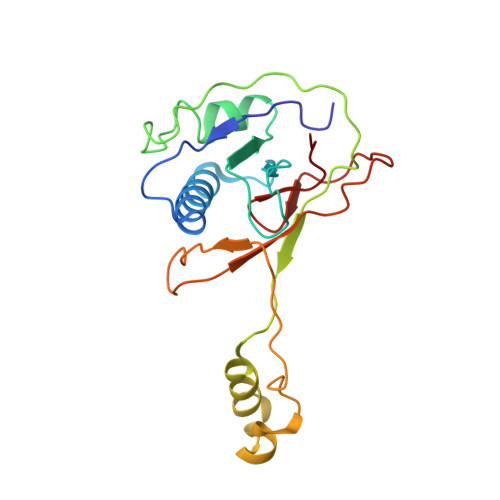
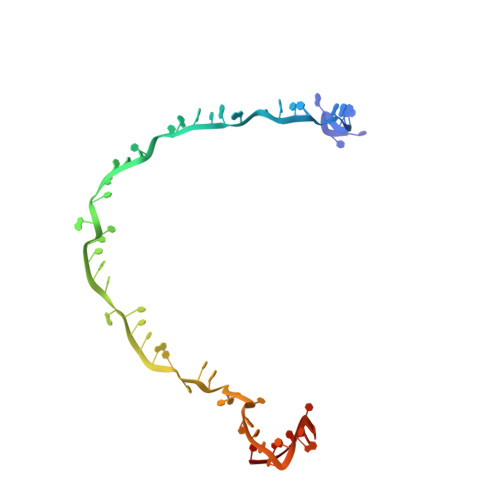
Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Rollins, M.F., Chowdhury, S., Carter, J., Golden, S.M., Miettinen, H.M., Santiago-Frangos, A., Faith, D., Lawrence, C.M., Lander, G.C., Wiedenheft, B.(2019) Mol Cell 74: 132-142.e5
- PubMed: 30872121
- DOI: https://doi.org/10.1016/j.molcel.2019.02.001
- Primary Citation of Related Structures:
6NE0 - PubMed Abstract:
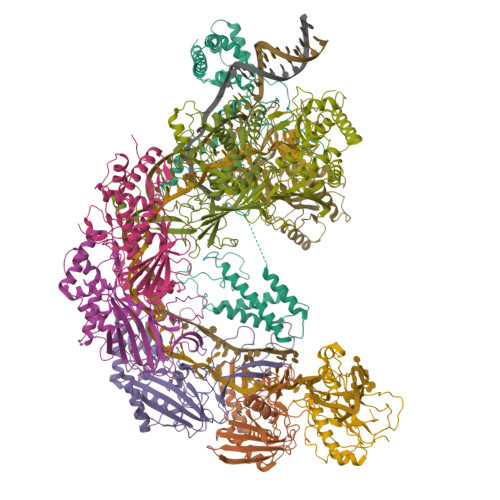
Bacteria and archaea have evolved sophisticated adaptive immune systems that rely on CRISPR RNA (crRNA)-guided detection and nuclease-mediated elimination of invading nucleic acids. Here, we present the cryo-electron microscopy (cryo-EM) structure of the type I-F crRNA-guided surveillance complex (Csy complex) from Pseudomonas aeruginosa bound to a double-stranded DNA target. Comparison of this structure to previously determined structures of this complex reveals a ∼180-degree rotation of the C-terminal helical bundle on the "large" Cas8f subunit. We show that the double-stranded DNA (dsDNA)-induced conformational change in Cas8f exposes a Cas2/3 "nuclease recruitment helix" that is structurally homologous to a virally encoded anti-CRISPR protein (AcrIF3). Structural homology between Cas8f and AcrIF3 suggests that AcrIF3 is a mimic of the Cas8f nuclease recruitment helix.
Organizational Affiliation:
Department of Microbiology and Immunology, Montana State University, Bozeman, MT 59717, USA.