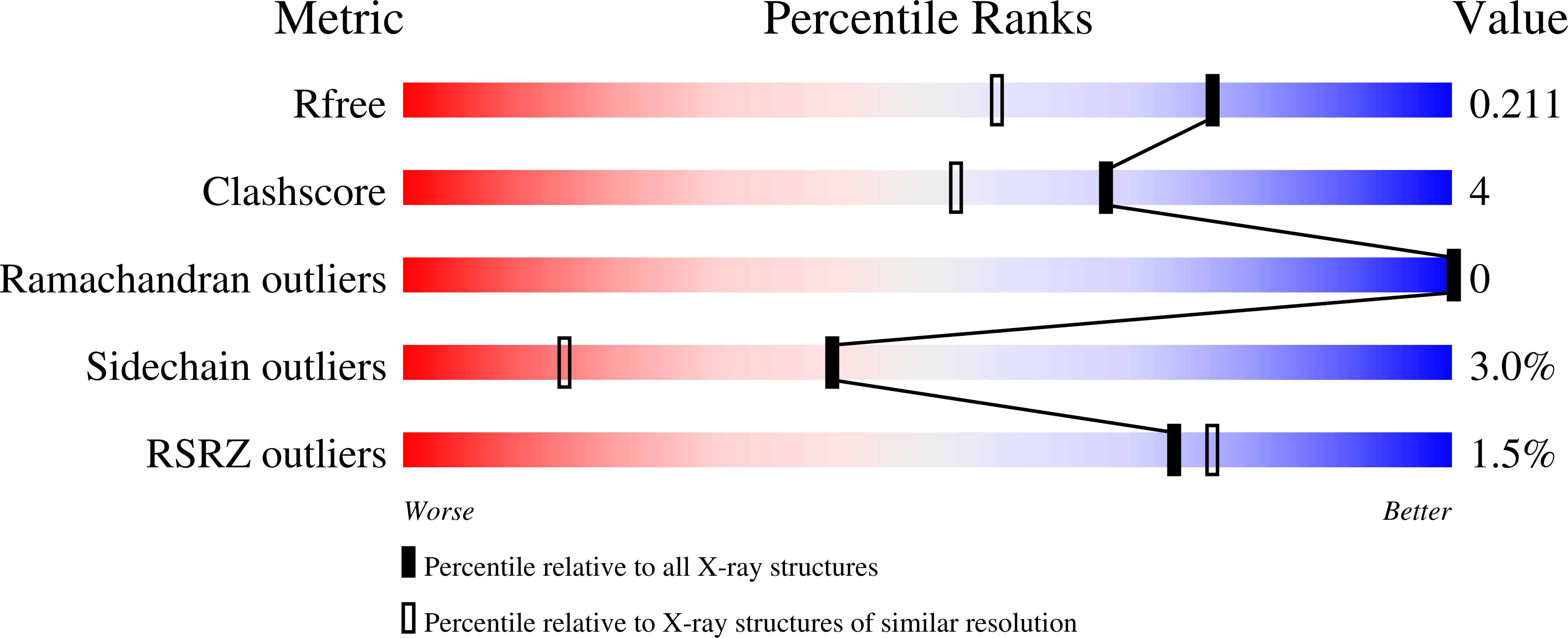
Crystal structure of human pyridoxine 5-phophate oxidase, R116Q variant
Mackinnon, S., Wilson, M.P., Shrestha, L., Bezerra, G.A., Newman, J., Fox, N., Sorrell, F., Arrowsmith, C.H., Edwards, A., Bountra, C., Clayton, P.T., Mills, P.B., Yue, W.W.To be published.