From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
Wang, Y., Cai, W., Tang, T., Liu, Q., Yang, T., Yang, L., Ma, Y., Zhang, G., Huang, Y., Song, X., Orband-Miller, L.A., Wu, Q., Zhou, L., Xiang, Z., Xiang, J.N., Leung, S., Shao, L., Lin, X., Lobera, M., Ren, F.(2018) ACS Med Chem Lett 9: 120-124
- PubMed: 29456799
- DOI: https://doi.org/10.1021/acsmedchemlett.7b00476
- Primary Citation of Related Structures:
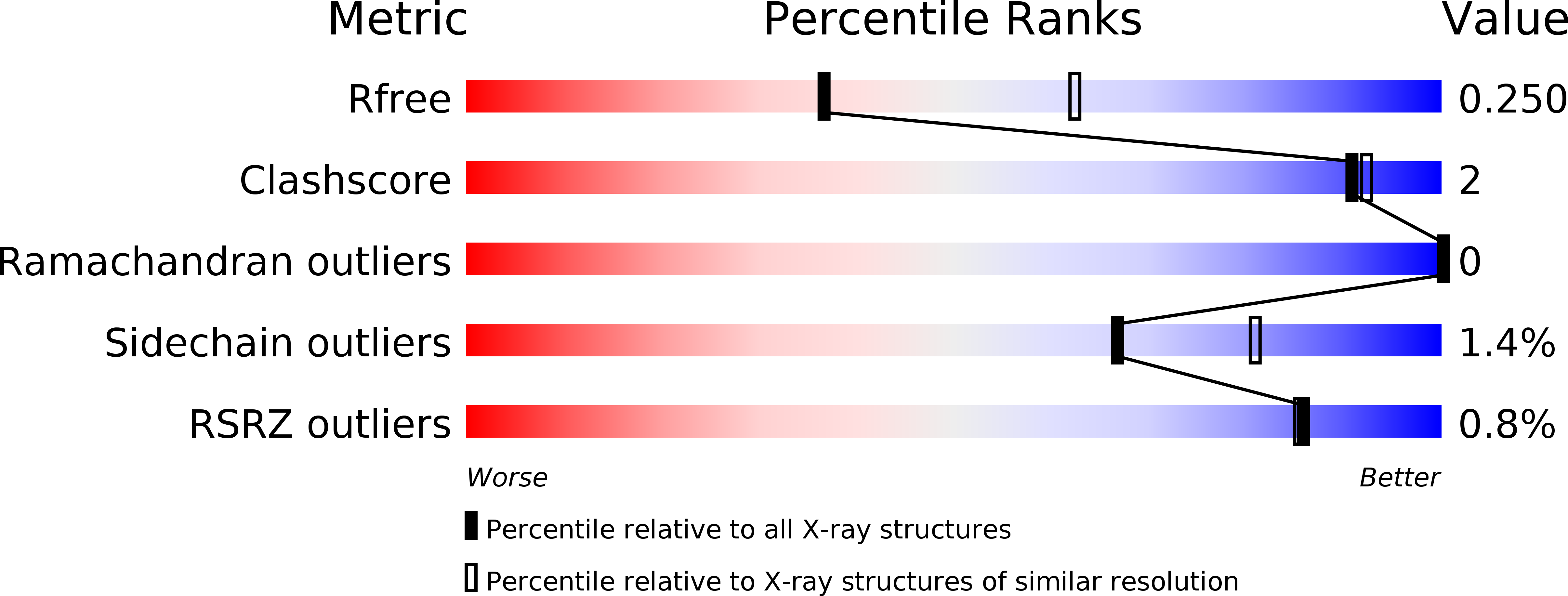
5YP5, 5YP6 - PubMed Abstract:
Biaryl amides as new RORγt modulators were discovered. The crystal structure of biaryl amide agonist 6 in complex with RORγt ligand binding domain (LBD) was resolved, and both "short" and "long" inverse agonists were obtained by removing from 6 or adding to 6 a proper structural moiety. While "short" inverse agonist ( 8 ) recruits a corepressor peptide and dispels a coactivator peptide, "long" inverse agonist ( 9 ) dispels both. The two types of inverse agonists can be utilized as potential tools to study mechanisms of Th17 transcriptional network inhibition and related disease biology.
Organizational Affiliation:
School of Pharmacy, Fudan University, 826 Zhangheng Road, Pudong, Shanghai 201203, China.