Structure-based optimization of 1H-imidazole-2-carboxamides as Axl kinase inhibitors utilizing a Mer mutant surrogate.
Keung, W., Boloor, A., Brown, J., Kiryanov, A., Gangloff, A., Lawson, J.D., Skene, R., Hoffman, I., Atienza, J., Kahana, J., De Jong, R., Farrell, P., Balakrishna, D., Halkowycz, P.(2017) Bioorg Med Chem Lett 27: 1099-1104
- PubMed: 28082036
- DOI: https://doi.org/10.1016/j.bmcl.2016.12.024
- Primary Citation of Related Structures:
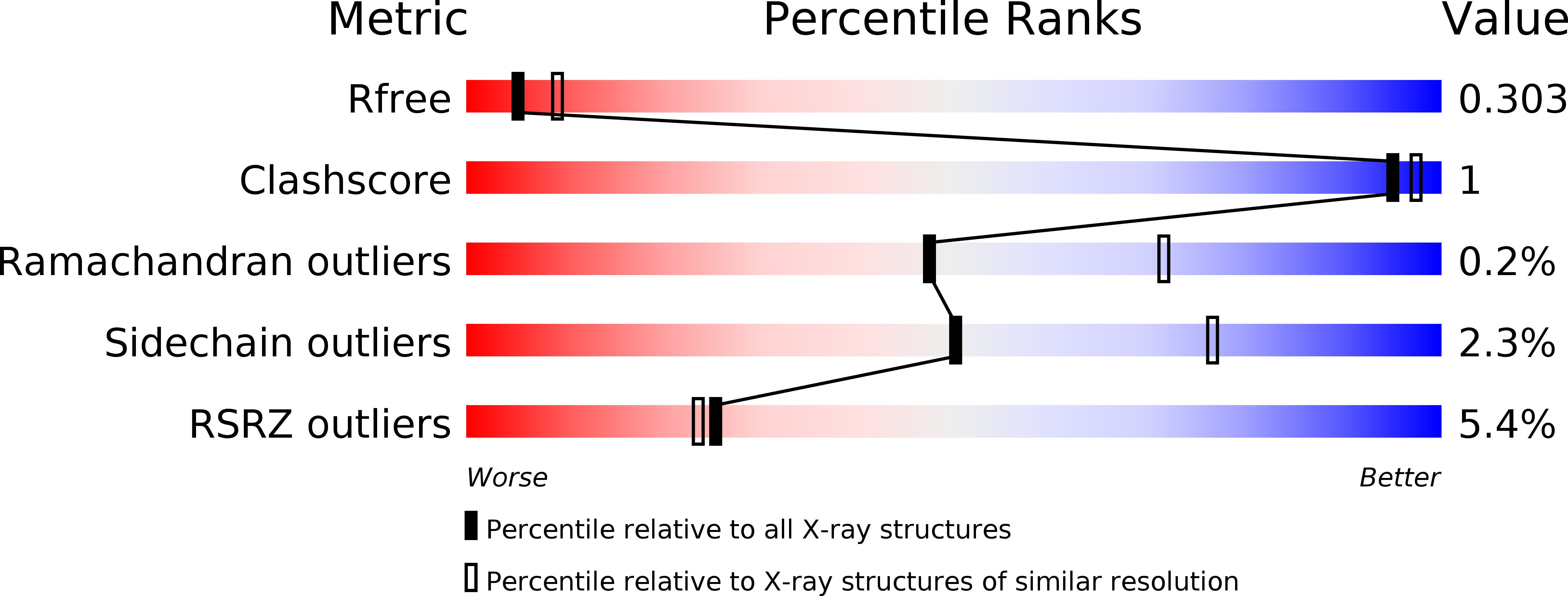
5TC0, 5TD2 - PubMed Abstract:
Axl has been a target of interest in the oncology field for several years based on its role in various oncogenic processes. To date, no wild-type Axl crystal structure has been reported. Herein, we describe the structure-based optimization of a novel chemotype of Axl inhibitors, 1H-imidazole-2-carboxamide, using a mutated kinase homolog, Mer(I650M), as a crystallographic surrogate. Iterative optimization of the initial lead compound (1) led to compound (21), a selective and potent inhibitor of wild-type Axl. Compound (21) will serve as a useful compound for further in vivo studies.
Organizational Affiliation:
Medicinal Chemistry, Takeda California, 10410 Science Center Drive, San Diego, CA 92121, United States. Electronic address: walter.keung@takeda.com.