2,8-Disubstituted-1,6-Naphthyridines and 4,6-Disubstituted-Isoquinolines with Potent, Selective Affinity for CDK8/19.
Mallinger, A., Schiemann, K., Rink, C., Sejberg, J., Honey, M.A., Czodrowski, P., Stubbs, M., Poeschke, O., Busch, M., Schneider, R., Schwarz, D., Musil, D., Burke, R., Urbahns, K., Workman, P., Wienke, D., Clarke, P.A., Raynaud, F.I., Eccles, S.A., Esdar, C., Rohdich, F., Blagg, J.(2016) ACS Med Chem Lett 7: 573-578
- PubMed: 27326329
- DOI: https://doi.org/10.1021/acsmedchemlett.6b00022
- Primary Citation of Related Structures:
5I5Z - PubMed Abstract:
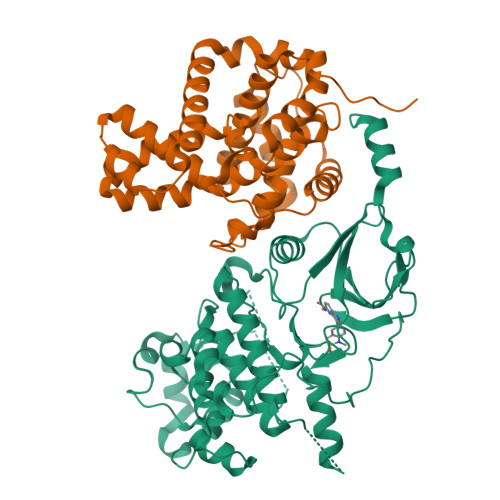
We demonstrate a designed scaffold-hop approach to the discovery of 2,8-disubstituted-1,6-naphthyridine- and 4,6-disubstituted-isoquinoline-based dual CDK8/19 ligands. Optimized compounds in both series exhibited rapid aldehyde oxidase-mediated metabolism, which could be abrogated by introduction of an amino substituent at C5 of the 1,6-naphthyridine scaffold or at C1 of the isoquinoline scaffold. Compounds 51 and 59 were progressed to in vivo pharmacokinetic studies, and 51 also demonstrated sustained inhibition of STAT1(SER727) phosphorylation, a biomarker of CDK8 inhibition, in an SW620 colorectal carcinoma human tumor xenograft model following oral dosing.
Organizational Affiliation:
Cancer Research UK Cancer Therapeutics Unit, The Institute of Cancer Research , London SW7 3RP, UK.