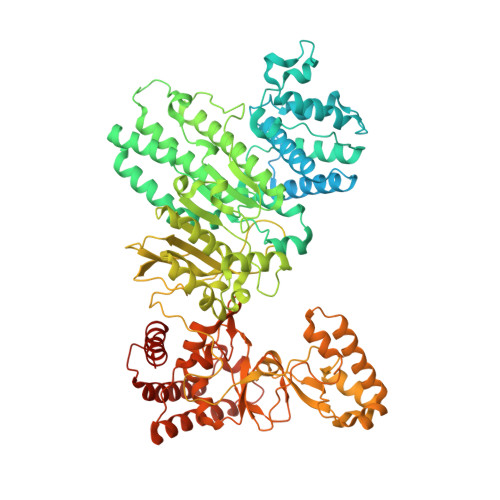
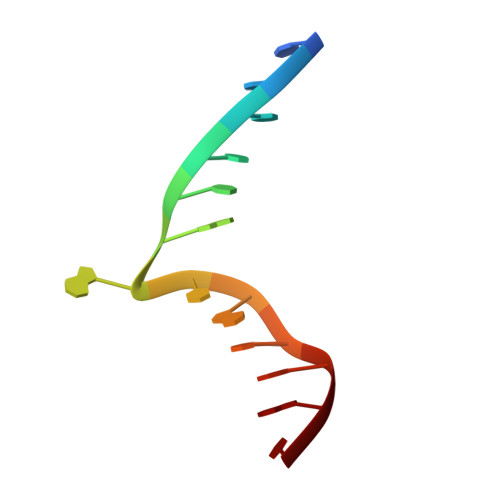
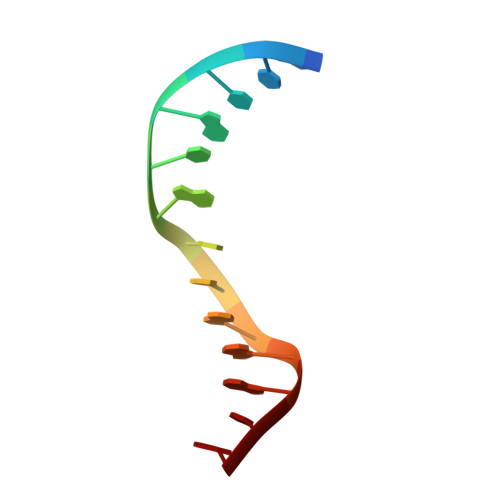
Structure of Type IIL Restriction-Modification Enzyme MmeI in Complex with DNA Has Implications for Engineering New Specificities.
Callahan, S.J., Luyten, Y.A., Gupta, Y.K., Wilson, G.G., Roberts, R.J., Morgan, R.D., Aggarwal, A.K.(2016) PLoS Biol 14: e1002442-e1002442
- PubMed: 27082731
- DOI: https://doi.org/10.1371/journal.pbio.1002442
- Primary Citation of Related Structures:
5HR4 - PubMed Abstract:
The creation of restriction enzymes with programmable DNA-binding and -cleavage specificities has long been a goal of modern biology. The recently discovered Type IIL MmeI family of restriction-and-modification (RM) enzymes that possess a shared target recognition domain provides a framework for engineering such new specificities. However, a lack of structural information on Type IIL enzymes has limited the repertoire that can be rationally engineered. We report here a crystal structure of MmeI in complex with its DNA substrate and an S-adenosylmethionine analog (Sinefungin). The structure uncovers for the first time the interactions that underlie MmeI-DNA recognition and methylation (5'-TCCRAC-3'; R = purine) and provides a molecular basis for changing specificity at four of the six base pairs of the recognition sequence (5'-TCCRAC-3'). Surprisingly, the enzyme is resilient to specificity changes at the first position of the recognition sequence (5'-TCCRAC-3'). Collectively, the structure provides a basis for engineering further derivatives of MmeI and delineates which base pairs of the recognition sequence are more amenable to alterations than others.
Organizational Affiliation:
Department of Structural and Chemical Biology, Mount Sinai School of Medicine, New York, New York, United States of America.