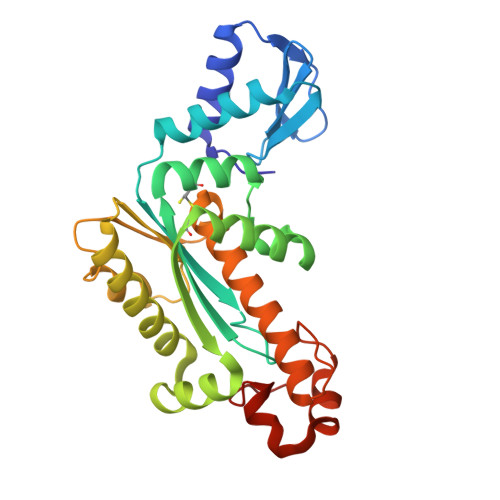
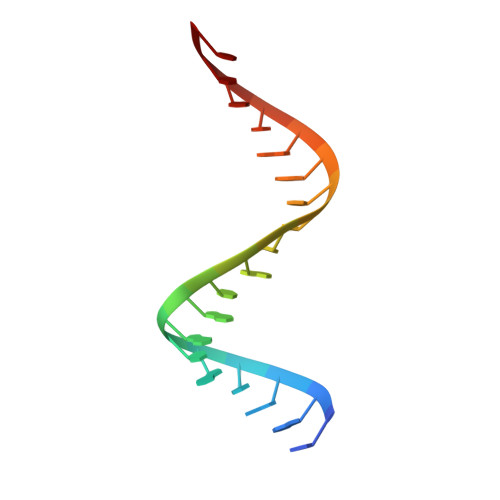
Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition.
Figiel, M., Nowotny, M.(2014) Nucleic Acids Res 42: 9285-9294
- PubMed: 25016521
- DOI: https://doi.org/10.1093/nar/gku615
- Primary Citation of Related Structures:
4PY5 - PubMed Abstract:
RNases H participate in the replication and maintenance of genomic DNA. RNase H1 cleaves the RNA strand of RNA/DNA hybrids, and RNase H2 in addition hydrolyzes the RNA residue of RNA-DNA junctions. RNase H3 is structurally closely related to RNases H2, but its biochemical properties are similar to type 1 enzymes. Its unique N-terminal substrate-binding domain (N-domain) is related to TATA-binding protein. Here, we report the first crystal structure of RNase H3 in complex with its RNA/DNA substrate. Just like RNases H1, type 3 enzyme recognizes the 2'-OH groups of the RNA strand and detects the DNA strand by binding a phosphate group and inducing B-form conformation. Moreover, the N-domain recognizes RNA and DNA in a manner that is highly similar to the hybrid-binding domain of RNases H1. Our structure demonstrates a remarkable example of parallel evolution of the elements used in the specific recognition of RNA and DNA.
Organizational Affiliation:
Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, 4 Trojdena Street, 02-109 Warsaw, Poland.