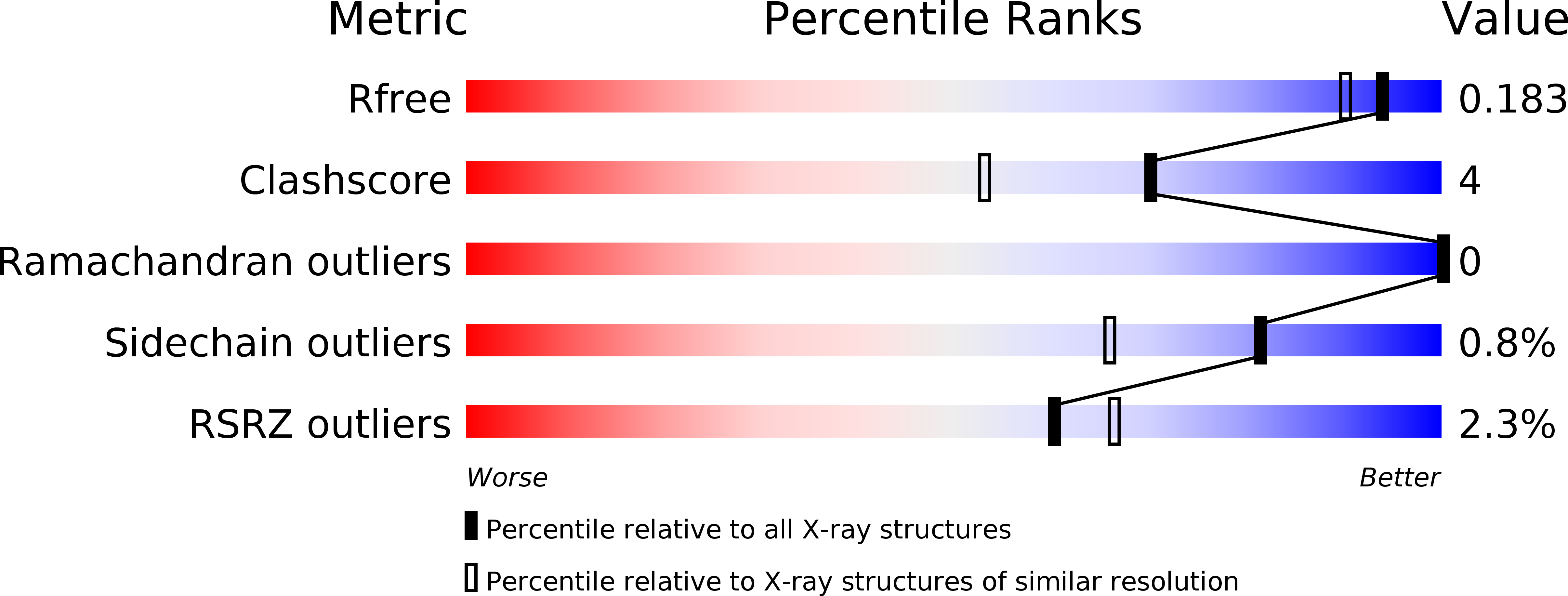
Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Sharifi, R., Morra, R., Appel, C.D., Tallis, M., Chioza, B., Jankevicius, G., Simpson, M.A., Matic, I., Ozkan, E., Golia, B., Schellenberg, M.J., Weston, R., Williams, J.G., Rossi, M.N., Galehdari, H., Krahn, J., Wan, A., Trembath, R.C., Crosby, A.H., Ahel, D., Hay, R., Ladurner, A.G., Timinszky, G., Williams, R.S., Ahel, I.(2013) EMBO J 32: 1225-1237
- PubMed: 23481255
- DOI: https://doi.org/10.1038/emboj.2013.51
- Primary Citation of Related Structures:
4J5Q, 4J5R, 4J5S - PubMed Abstract:
Adenosine diphosphate (ADP)-ribosylation is a post-translational protein modification implicated in the regulation of a range of cellular processes. A family of proteins that catalyse ADP-ribosylation reactions are the poly(ADP-ribose) (PAR) polymerases (PARPs). PARPs covalently attach an ADP-ribose nucleotide to target proteins and some PARP family members can subsequently add additional ADP-ribose units to generate a PAR chain. The hydrolysis of PAR chains is catalysed by PAR glycohydrolase (PARG). PARG is unable to cleave the mono(ADP-ribose) unit directly linked to the protein and although the enzymatic activity that catalyses this reaction has been detected in mammalian cell extracts, the protein(s) responsible remain unknown. Here, we report the homozygous mutation of the c6orf130 gene in patients with severe neurodegeneration, and identify C6orf130 as a PARP-interacting protein that removes mono(ADP-ribosyl)ation on glutamate amino acid residues in PARP-modified proteins. X-ray structures and biochemical analysis of C6orf130 suggest a mechanism of catalytic reversal involving a transient C6orf130 lysyl-(ADP-ribose) intermediate. Furthermore, depletion of C6orf130 protein in cells leads to proliferation and DNA repair defects. Collectively, our data suggest that C6orf130 enzymatic activity has a role in the turnover and recycling of protein ADP-ribosylation, and we have implicated the importance of this protein in supporting normal cellular function in humans.
Organizational Affiliation:
Biomedical Sciences Division, Human Genetics Research Centre, St George's University of London, London, UK. [email protected]