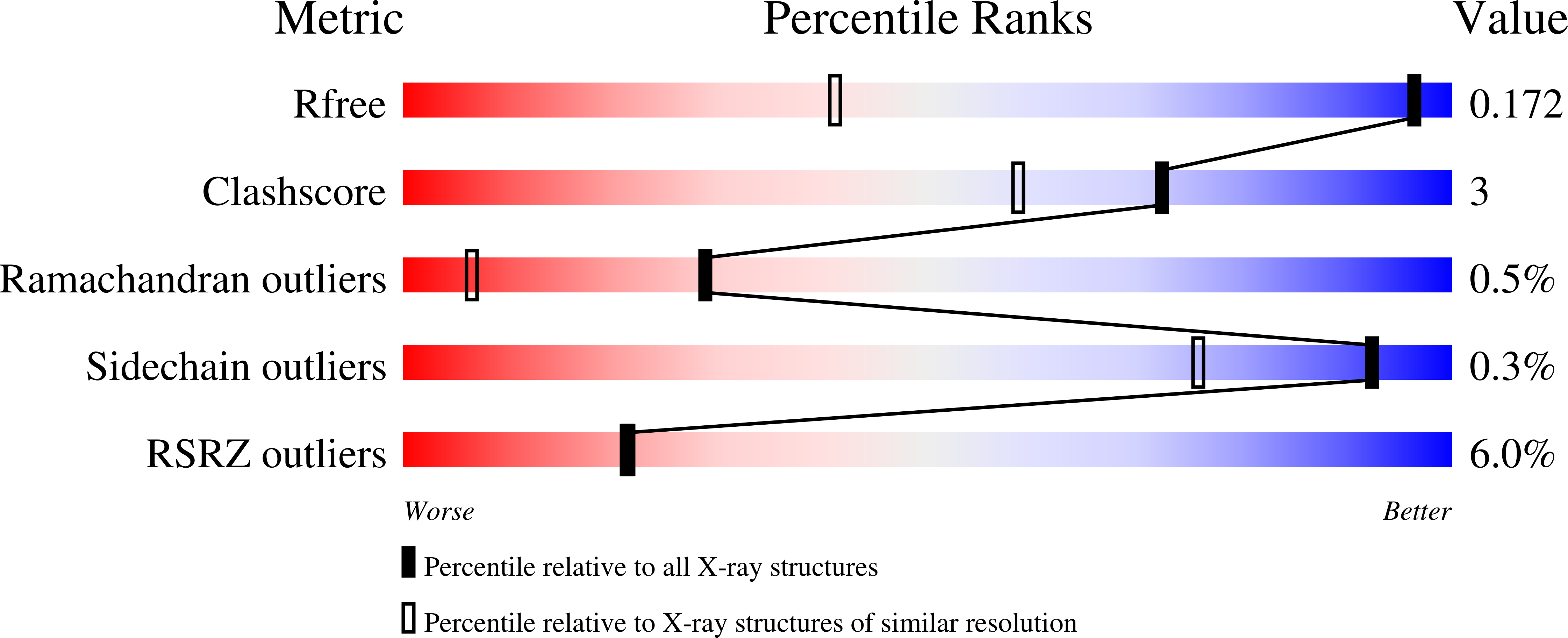
Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate
Vetting, M.W., Sauder, J.M., Morisco, L.L., Wasserman, S.R., Sojitra, S., Imker, H.J., Burley, S.K., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.