Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Liang, G., Chen, X., Aldous, S., Pu, S.F., Mehdi, S., Powers, E., Giovanni, A., Kongsamut, S., Xia, T., Zhang, Y., Wang, R., Gao, Z., Merriman, G., McLean, L.R., Morize, I.(2012) ACS Med Chem Lett 3: 159-164
- PubMed: 24900446
- DOI: https://doi.org/10.1021/ml200291e
- Primary Citation of Related Structures:
3VFE - PubMed Abstract:
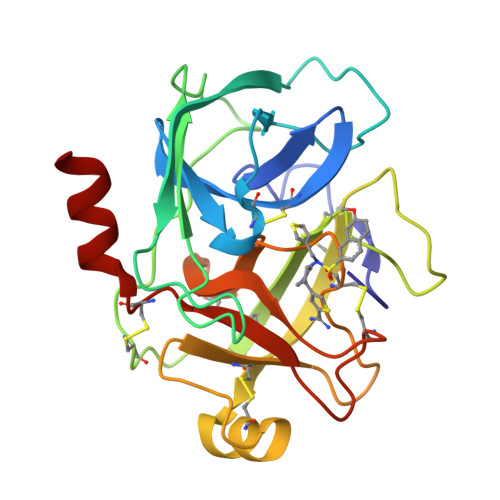
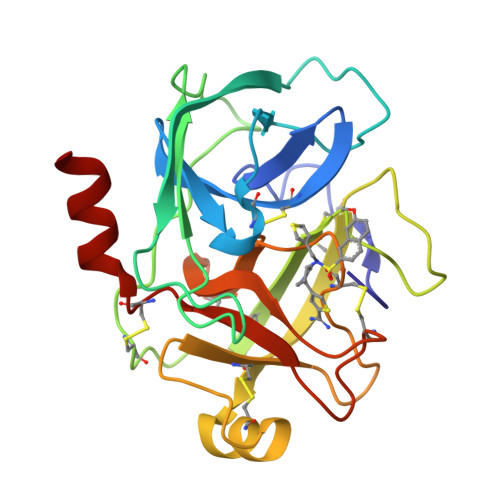
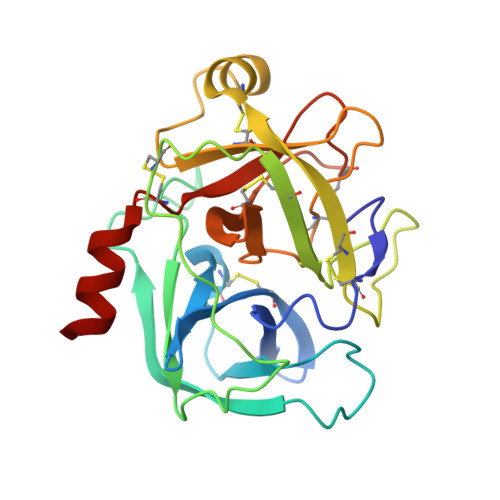
A series of compounds with an amidinothiophene P1 group and a pyrrolidinone-sulphonamide scaffold linker was identified as potent inhibitors of human kallikrein 6 by structure-based virtual screening based on the union accessible binding space of serine proteases. As the first series of potent nonmechanism-based hK6 inhibitors, they may be used as tool compounds for target validation. An X-ray structure of a representative compound complexed with hK6, resolved at a resolution of 1.88 Å, revealed that the amidinothiophene moiety bound in the S1 pocket and the pyrrolidinone-sulphonamide linker projected the aromatic tail into the S' pocket.
Organizational Affiliation:
Molecular Innovative Therapeutics, Fibrosis and Wound Repair, Global Pharmacovigilance & Epidemiology, Immunology and Inflammation Unit, Early to Candidate Unit, Aging Therapeutic Unit, and Biologics, Sanofi Pharmaceuticals , 1041 Route 202/206, Mailstop 203A, Bridgewater, New Jersey 08807, United States.