Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
Hermann, T., Dibrov, S., Ding, K., Brunn, N., Parker, M., Bergdahl, M., Wyles, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3' | 20 | Hepacivirus hominis | 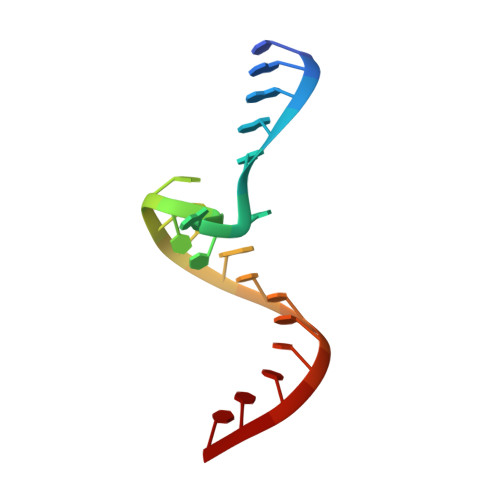 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3' | 16 | Hepacivirus hominis | 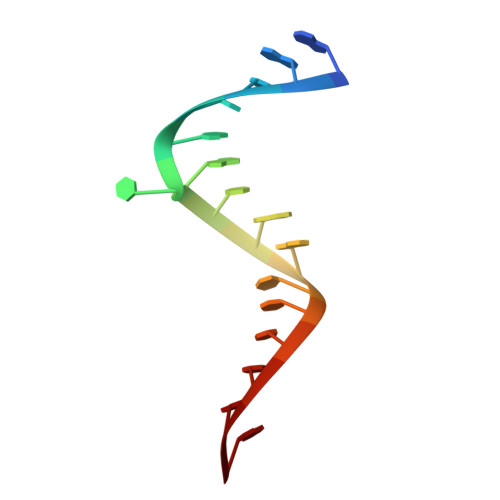 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SS0 Query on SS0 | K [auth A] | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine C18 H29 N5 O HTUXJUVSTFSWOD-CYBMUJFWSA-N |  | ||
| SO4 Query on SO4 | H [auth A], I [auth A], J [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| MG Query on MG | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 32.054 | α = 90 |
| b = 33.582 | β = 90 |
| c = 81.046 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |