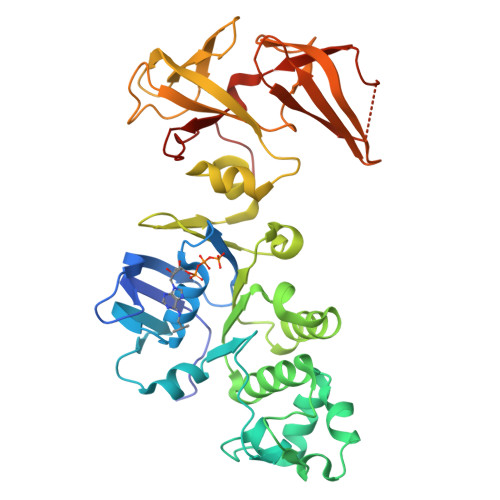
Entity ID: 1 |
|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
|---|
| Fusion complex of Cystic fibrosis transmembrane conductance regulator, residues 1193-1427 and Maltose/maltodextrin import ATP-binding protein malK, residues 219-371 | | 390 | Homo sapiens, Escherichia coli K-12
This entity is chimeric | Mutation(s): 7
Gene Names: CFTR, malK
EC: 5.6.1.6 (UniProt), 7.5.2.1 (UniProt)
|  |
UniProt & NIH Common Fund Data Resources |
Find proteins for P13569 (Homo sapiens) |
|
Find proteins for P68187 (Escherichia coli (strain K12)) |
Entity Groups
|
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity |
| UniProt Groups | P13569P68187 |
Sequence Annotations |
|