Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents.
Abad-Zapatero, C., Goldman, R., Muchmore, S.W., Hutchins, C., Stewart, K., Navaza, J., Payne, C.D., Ray, T.L.(1996) Protein Sci 5: 640-652
- PubMed: 8845753
- DOI: https://doi.org/10.1002/pro.5560050408
- Primary Citation of Related Structures:
1ZAP - PubMed Abstract:
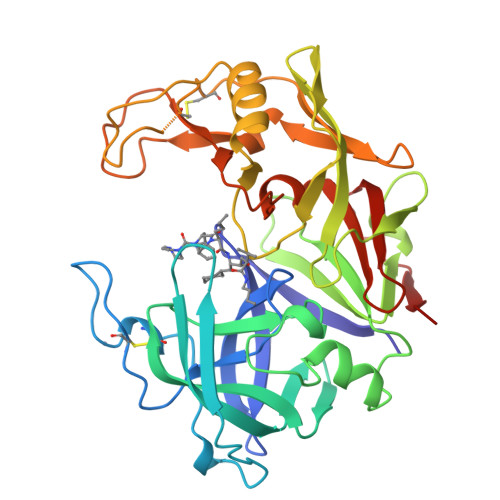
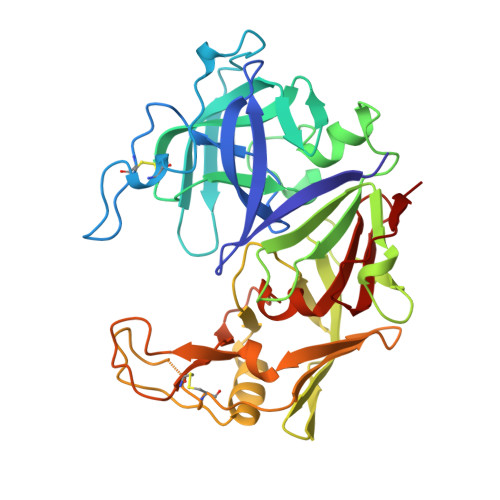
The three-dimensional structure of a secreted aspartic protease from Candida albicans complexed with a potent inhibitor reveals variations on the classical aspartic protease theme that dramatically alter the specificity of this class of enzymes. The structure presents: (1) an 8-residue insertion near the first disulfide (Cys 45-Cys 50, pepsin numbering) that results in a broad flap extending toward the active site; (2) a 7-residue deletion replacing helix hN2 (Ser 110-Tyr 114), which enlarges the S3 pocket; (3) a short polar connection between the two rigid body domains that alters their relative orientation and provides certain specificity; and (4) an ordered 11-residue addition at the carboxy terminus. The inhibitor binds in an extended conformation and presents a branched structure at the P3 position. The implications of these findings for the design of potent antifungal agents are discussed.
Organizational Affiliation:
Laboratory of Protein Crystallography, Abbott Laboratories, Abbott Park, Illinois 60064-3500, USA.