Crystal structure of beta-xylosidase from clostridium acetobutylicum
Teplyakov, A., Fedorov, E., Gilliland, G.L., Almo, S.C., New York Structural Genomics Research Consortium (NYSGRC)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-xylosidase, family 43 glycosyl hydrolase | 542 | Clostridium acetobutylicum ATCC 824 | Mutation(s): 4 Gene Names: CAC3452 (xynD) EC: 3.2.1.37 | 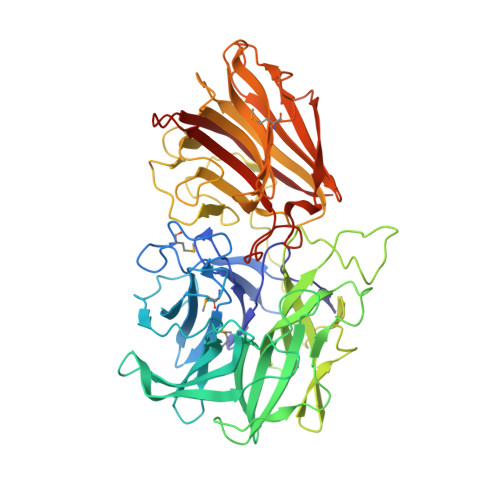 | |
UniProt | |||||
Find proteins for Q97DM1 (Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / IAM 19013 / LMG 5710 / NBRC 13948 / NRRL B-527 / VKM B-1787 / 2291 / W)) Explore Q97DM1 Go to UniProtKB: Q97DM1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q97DM1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | H [auth A], HA [auth D], I [auth A], Y [auth C] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | F [auth A] FA [auth D] G [auth A] GA [auth D] P [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | AA [auth C] BA [auth C] CA [auth C] DA [auth C] IA [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth A], EA [auth D], O [auth B], W [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 89.13 | α = 90 |
| b = 143.73 | β = 90 |
| c = 188.69 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| AMoRE | phasing |