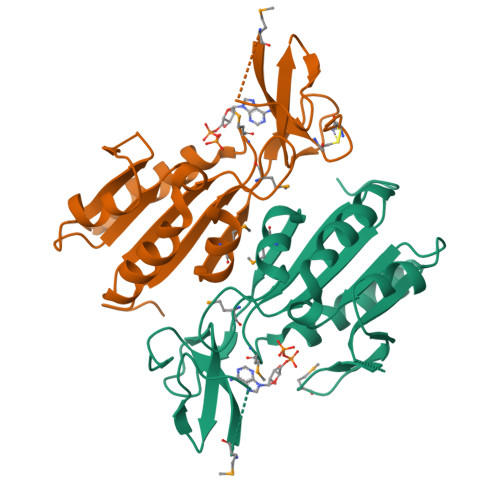
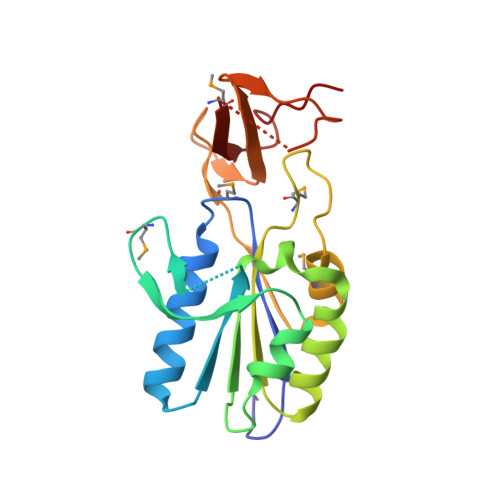
X-ray structure of Clostridium acetobutylicum thymidine kinase with ADP. Northeast Structural Genomics Target CAR26.
Kuzin, A.P., Abashidze, M., Forouhar, F., Vorobiev, S.M., Acton, T.B., Ma, L.-C., Xiao, R., Montelione, G.T., Tong, L., Hunt, J.F.To be published.