Full Text |
QUERY: Chemical ID(s) = 35G | MyPDB Login | Search API |
| Search Summary | This query matches 7 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 7 of 7 Structures Page 1 of 1 Sort by
NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINALadner, J.E., Abdulaev, N.G., Kakuev, D.L., Karaschuk, G.N., Tordova, M., Eisenstein, E., Fujiwara, J.H., Ridge, K.D., Gilliland, G.L. (1999) Acta Crystallogr D Biol Crystallogr 55: 1127-1135
Solution Structure of cGMP-binding GAF domain of Phosphodiesterase 5Heikaus, C.C., Stout, J.R., Sekharan, M.R., Eakin, C.M., Rajagopal, P., Brzovic, P.S., Beavo, J.A., Klevit, R.E. (2008) J Biological Chem 283: 22749-22759
crystal structure of PDE10A2 mutant D674A in complex with cGMPWang, H.C., Liu, Y.D., Hou, J., Zheng, M.Y., Robinson, H. (2007) Proc Natl Acad Sci U S A 104: 5782-5787
Crystal structure of the cGMP-bound GAF a domain from the photoreceptor phosphodiesterase 6CMartinez, S.E., Heikaus, C.C., Klevit, R.E., Beavo, J.A. (2008) J Biological Chem 283: 25913-25919
PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMPCampbell, J.C., Reger, A.S., Huang, G.Y., Sankaran, B., Kim, J.J., Kim, C.W. (2016) J Biological Chem 291: 5623-5633
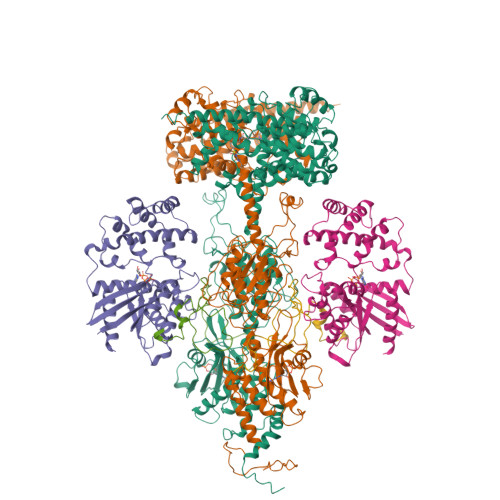
Cryo-EM structure of phosphodiesterase 6(2019) Sci Adv 5: eaav4322-eaav4322
Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6Gao, Y., Eskici, G., Ramachandran, S., Skiniotis, G., Cerione, R.A. (2020) Mol Cell 80: 237
1 to 7 of 7 Structures Page 1 of 1 Sort by |