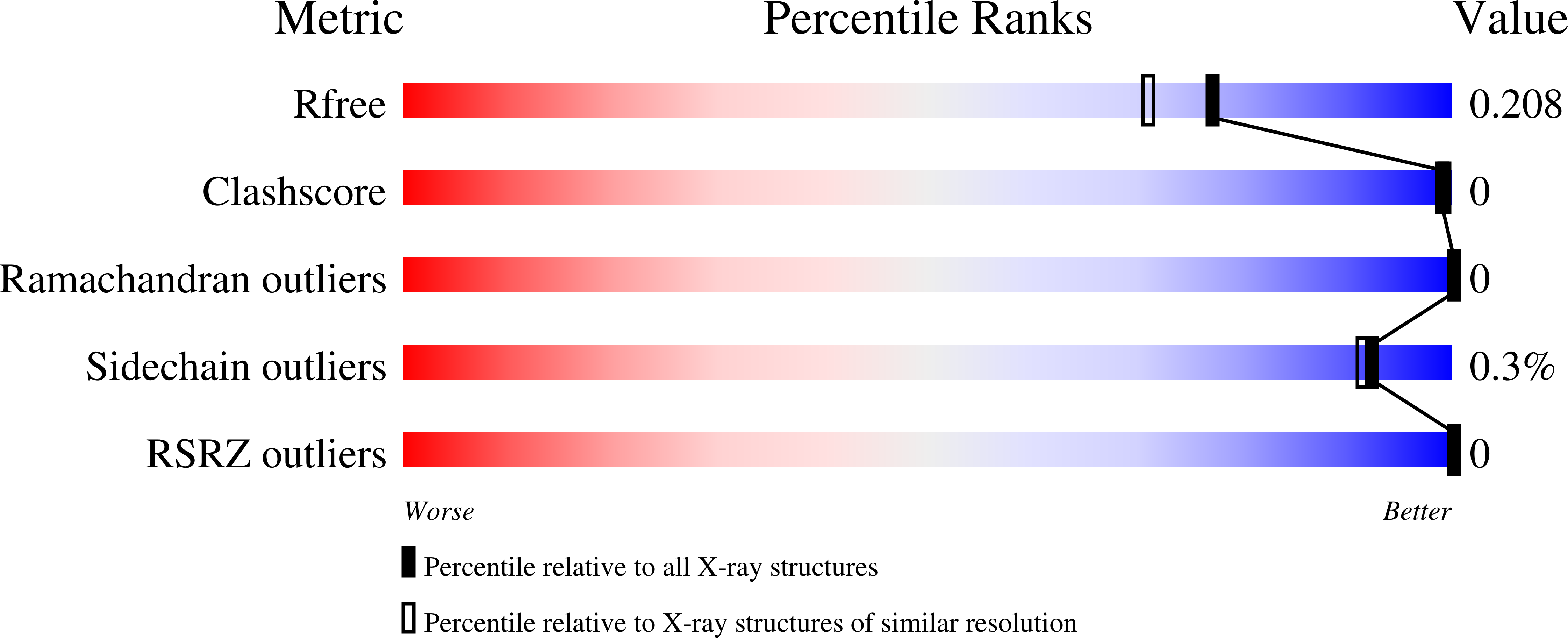
Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Fowler, C.A., Hemsworth, G.R., Cuskin, F., Hart, S., Turkenburg, J., Gilbert, H.J., Walton, P.H., Davies, G.J.(2018) Acta Crystallogr D Struct Biol 74: 946-955
- PubMed: 30289404
- DOI: https://doi.org/10.1107/S2059798318009737
- Primary Citation of Related Structures:
6G00, 6G09, 6G0B, 6G0N - PubMed Abstract:
The biological conversion of lignocellulosic matter into high-value chemicals or biofuels is of increasing industrial importance as the sector slowly transitions away from nonrenewable sources. Many industrial processes involve the use of cellulolytic enzyme cocktails - a selection of glycoside hydrolases and, increasingly, polysaccharide oxygenases - to break down recalcitrant plant polysaccharides. ORFs from the genome of Teredinibacter turnerae, a symbiont hosted within the gills of marine shipworms, were identified in order to search for enzymes with desirable traits. Here, a putative T. turnerae glycoside hydrolase from family 8, hereafter referred to as TtGH8, is analysed. The enzyme is shown to be active against β-1,4-xylan and mixed-linkage (β-1,3,β-1,4) marine xylan. Kinetic parameters, obtained using high-performance anion-exchange chromatography with pulsed amperometric detection and 3,5-dinitrosalicyclic acid reducing-sugar assays, show that TtGH8 catalyses the hydrolysis of β-1,4-xylohexaose with a k cat /K m of 7.5 × 10 7 M -1 min -1 but displays maximal activity against mixed-linkage polymeric xylans, hinting at a primary role in the degradation of marine polysaccharides. The three-dimensional structure of TtGH8 was solved in uncomplexed and xylobiose-, xylotriose- and xylohexaose-bound forms at approximately 1.5 Å resolution; the latter was consistent with the greater k cat /K m for hexasaccharide substrates. A 2,5 B boat conformation observed in the -1 position of bound xylotriose is consistent with the proposed conformational itinerary for this class of enzyme. This work shows TtGH8 to be effective at the degradation of xylan-based substrates, notably marine xylan, further exemplifying the potential of T. turnerae for effective and diverse biomass degradation.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, The University of York, York YO10 5DD, England.