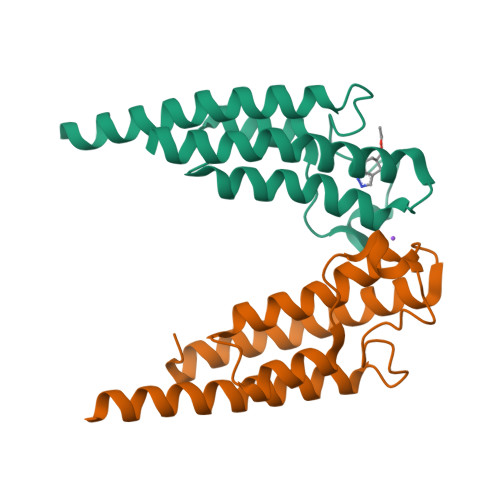
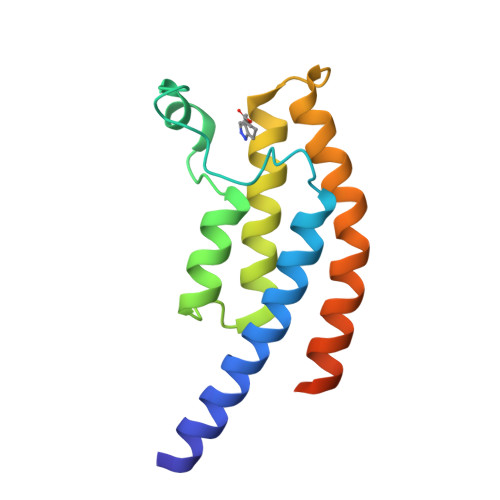
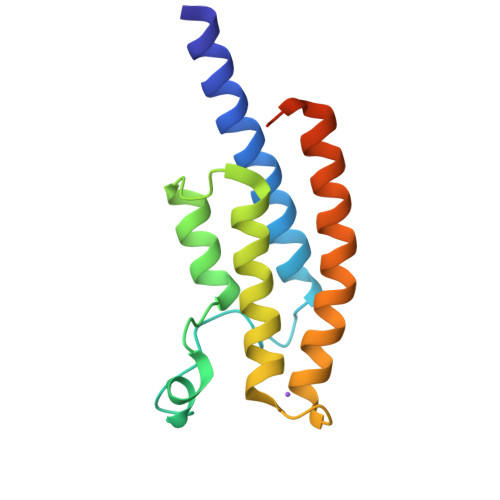
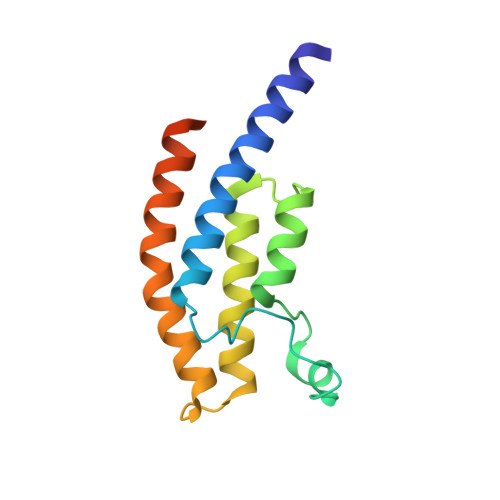
Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
Pearce, N.M., Fairhead, M., Strain-Damerell, C., Talon, R., Wright, N., Ng, J.T., Bradley, A., Cox, O., Bowkett, D., Collins, P., Brandao-Neto, J., Douangamath, A., Krojer, T., Burgess-Brown, N., Brennan, P., Arrowsmith, C.H., Edwards, E., Bountra, C., von Delft, F.To be published.