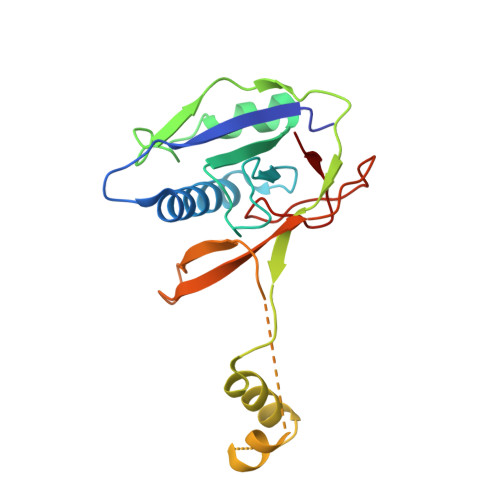
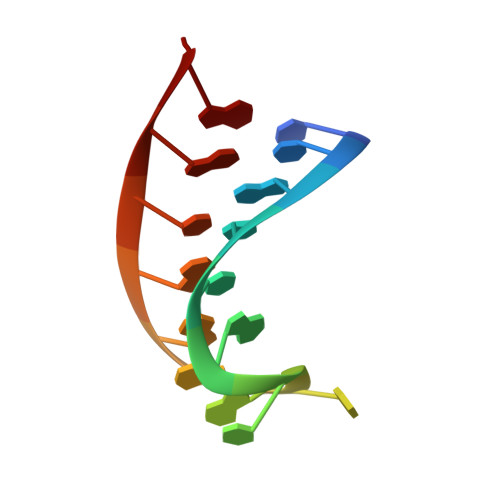
Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Haurwitz, R.E., Jinek, M., Wiedenheft, B., Zhou, K., Doudna, J.A.(2010) Science 329: 1355
- PubMed: 20829488
- DOI: https://doi.org/10.1126/science.1192272
- Primary Citation of Related Structures:
2XLI, 2XLJ, 2XLK - PubMed Abstract:
Many bacteria and archaea contain clustered regularly interspaced short palindromic repeats (CRISPRs) that confer resistance to invasive genetic elements. Central to this immune system is the production of CRISPR-derived RNAs (crRNAs) after transcription of the CRISPR locus. Here, we identify the endoribonuclease (Csy4) responsible for CRISPR transcript (pre-crRNA) processing in Pseudomonas aeruginosa. A 1.8 angstrom crystal structure of Csy4 bound to its cognate RNA reveals that Csy4 makes sequence-specific interactions in the major groove of the crRNA repeat stem-loop. Together with electrostatic contacts to the phosphate backbone, these enable Csy4 to bind selectively and cleave pre-crRNAs using phylogenetically conserved serine and histidine residues in the active site. The RNA recognition mechanism identified here explains sequence- and structure-specific processing by a large family of CRISPR-specific endoribonucleases.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.