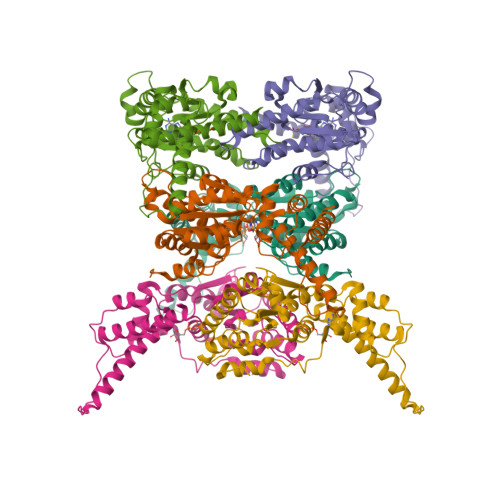
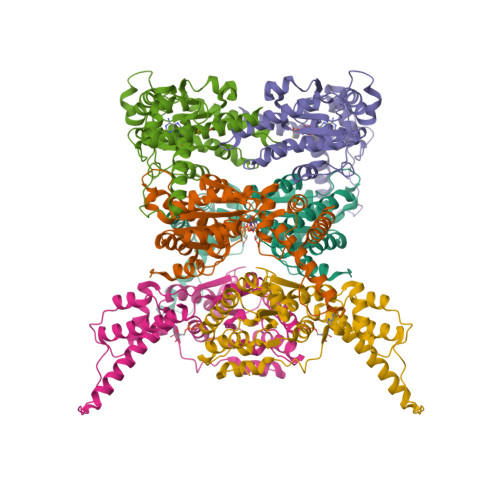
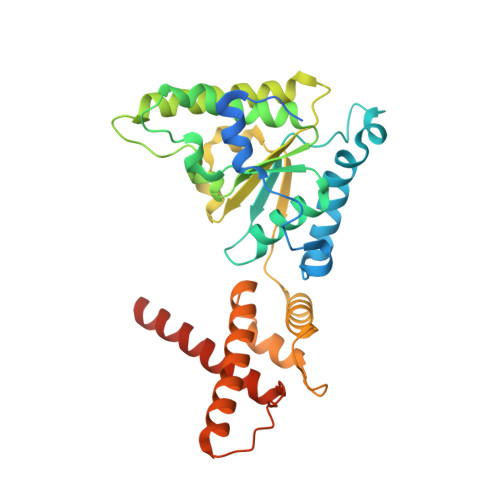
Cryo-EM Structure of AAA + ATPase Thorase Reveals Novel Helical Filament Formation.
Dar, M.A., Louder, R., Cortes, M., Chen, R., Ma, Q., Chakrabarti, M., Umanah, G.K.E., Dawson, T.M., Dawson, V.L.(2024) bioRxiv
- PubMed: 39605435
- DOI: https://doi.org/10.1101/2024.11.22.624887
- Primary Citation of Related Structures:
8VXT - PubMed Abstract:
The AAA+ (ATPases associated with a variety of cellular activities) ATPase, Thorase, also known as ATAD1, plays multiple roles in synaptic plasticity, mitochondrial quality control and mTOR signaling through disassembling protein complexes like AMPAR and mTORC1 in an ATP-dependent manner. The Oligomerization of Thorase is crucial for its disassembly and remodeling functions. We show that wild-type Thorase forms long helical filaments in vitro , dependent on ATP binding but not hydrolysis. We report the Cryogenic Electron Microscopy (cryo-EM) structure of the Thorase filament at a resolution of 4 Å, revealing the dimeric arrangement of the basic repeating unit that is formed through a distinct interface compared to the hexameric MSP1/ATAD1E193Q assembly. Structure-guided mutagenesis confirms the role of critical amino acid residues required for filament formation, oligomerization and disassembly of mTORC1 protein complex. Together, our data reveals a novel filament structure of Thorase and provides critical information that elucidates the mechanism underlying Thorase filament formation and Thorase-mediated disassembly of the mTORC1 complex.
Organizational Affiliation:
Neuroregeneration and Stem Cell Programs, Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, Maryland, United States of America.