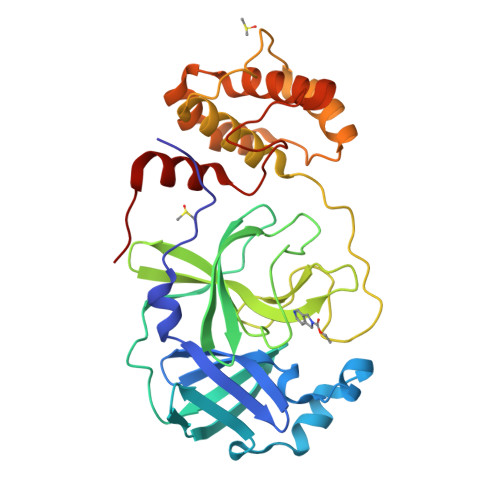
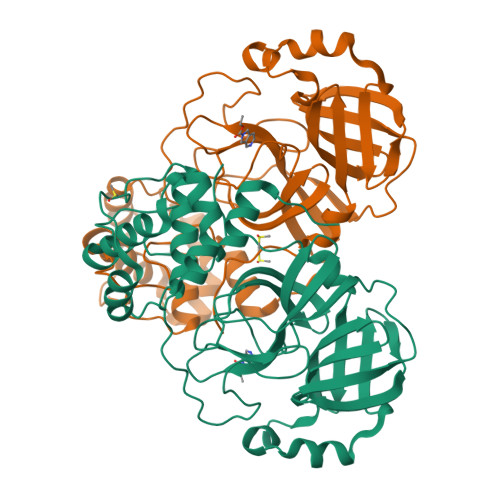
Orthorhombic SARS-CoV-2 main protease crystals provide higher success rate in fragment screening
Barthel, T., Wollenhaupt, J., Benz, L.S., Reincke, P., Zhang, L., Lennartz, F., Tabermann, H., Mueller, U., Meente, A., Hilgenfeld, R., Weiss, M.S.To be published.