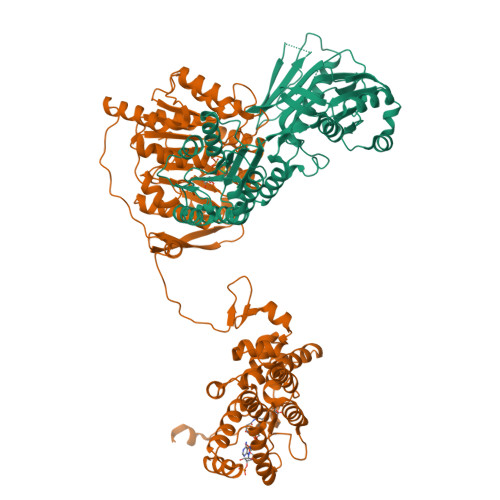
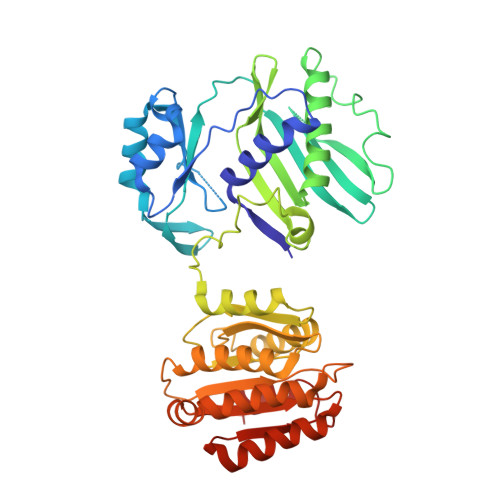
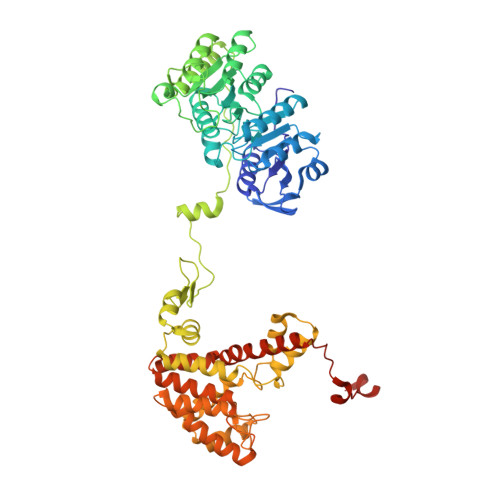
Acetyl-CoA is produced by the citrate synthase homology module of ATP-citrate lyase.
Verstraete, K., Verschueren, K.H.G., Dansercoer, A., Savvides, S.N.(2021) Nat Struct Mol Biol 28: 636-638
- PubMed: 34294920
- DOI: https://doi.org/10.1038/s41594-021-00624-3
- Primary Citation of Related Structures:
6Z2H, 6ZNW
Organizational Affiliation:
VIB-UGent Center for Inflammation Research, Ghent, Belgium. Kenneth.Verstraete@ugent.be.