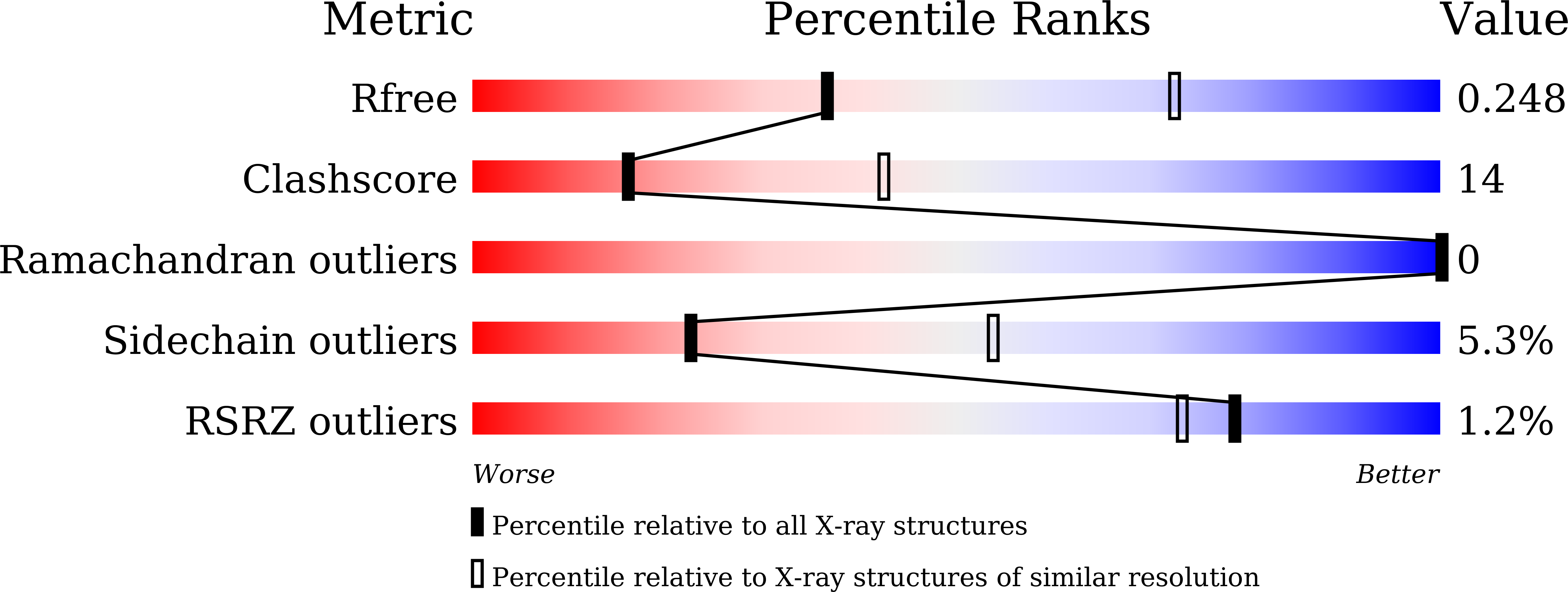
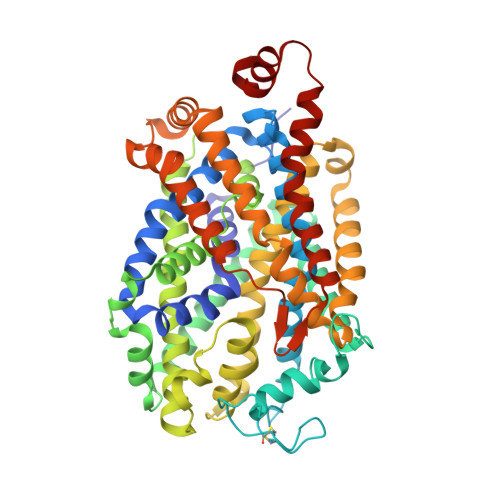
Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Pidathala, S., Mallela, A.K., Joseph, D., Penmatsa, A.(2021) Nat Commun 12: 2199-2199
- PubMed: 33850134
- DOI: https://doi.org/10.1038/s41467-021-22385-9
- Primary Citation of Related Structures:
6M0F, 6M0Z, 6M2R, 6M38, 6M3Z, 6M47 - PubMed Abstract:
Norepinephrine is a biogenic amine neurotransmitter that has widespread effects on alertness, arousal and pain sensation. Consequently, blockers of norepinephrine uptake have served as vital tools to treat depression and chronic pain. Here, we employ the Drosophila melanogaster dopamine transporter as a surrogate for the norepinephrine transporter and determine X-ray structures of the transporter in its substrate-free and norepinephrine-bound forms. We also report structures of the transporter in complex with inhibitors of chronic pain including duloxetine, milnacipran and a synthetic opioid, tramadol. When compared to dopamine, we observe that norepinephrine binds in a different pose, in the vicinity of subsite C within the primary binding site. Our experiments reveal that this region is the binding site for chronic pain inhibitors and a determinant for norepinephrine-specific reuptake inhibition, thereby providing a paradigm for the design of specific inhibitors for catecholamine neurotransmitter transporters.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.