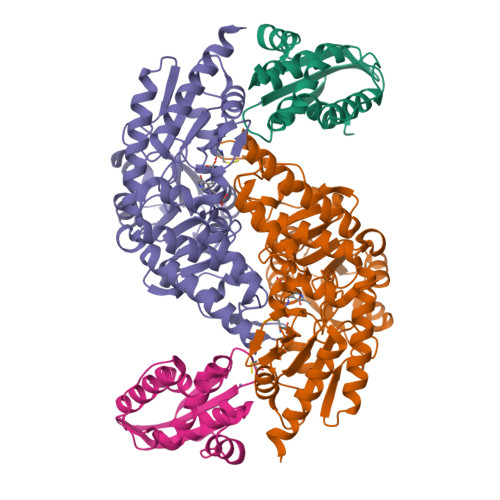
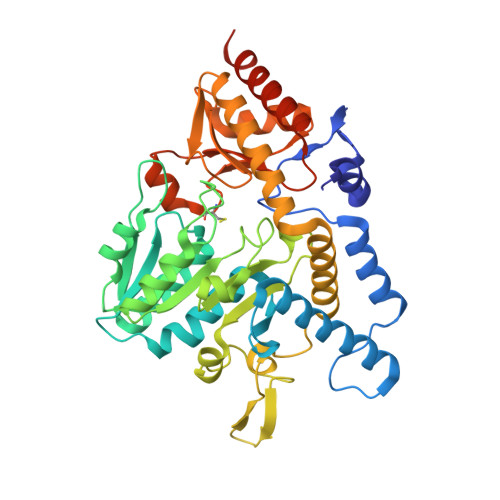
Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
Fujishiro, T., Terahata, T., Kunichika, K., Yokoyama, N., Maruyama, C., Asai, K., Takahashi, Y.(2017) J Am Chem Soc 139: 18464-18467
- PubMed: 29235855
- DOI: https://doi.org/10.1021/jacs.7b11307
- Primary Citation of Related Structures:
5XT5, 5XT6 - PubMed Abstract:
SufU is a zinc-containing protein involved in mobilization of sulfur from SufS for iron-sulfur cluster biogenesis of Bacillus subtilis. Structural basis for the sulfur transfer in SufS-SufU complex was revealed. A zinc-ligand exchange reaction upon SufS-SufU complexation provides a free thiol from Cys41 of SufU as a sulfur acceptor.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Graduate School of Science and Engineering, Saitama University , Shimo-ohkubo 255, Sakura-ku, Saitama 338-8570, Japan.