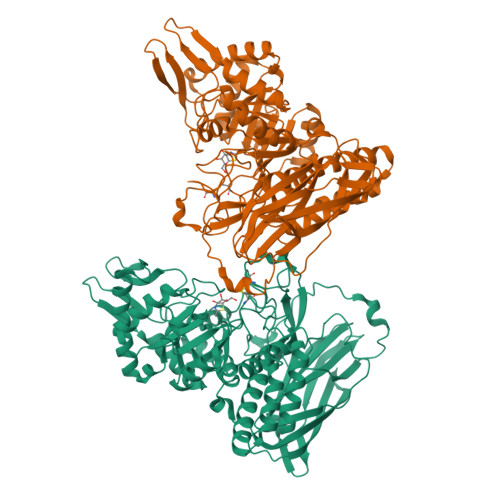
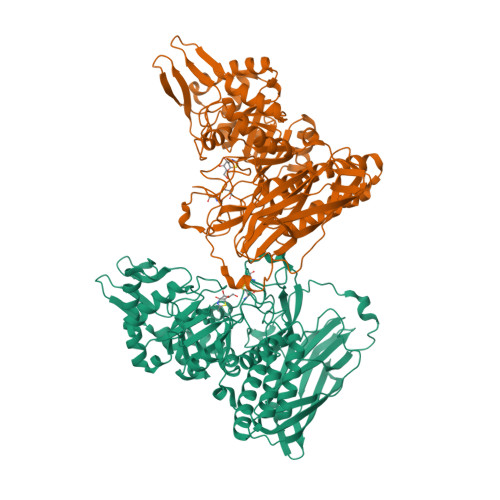
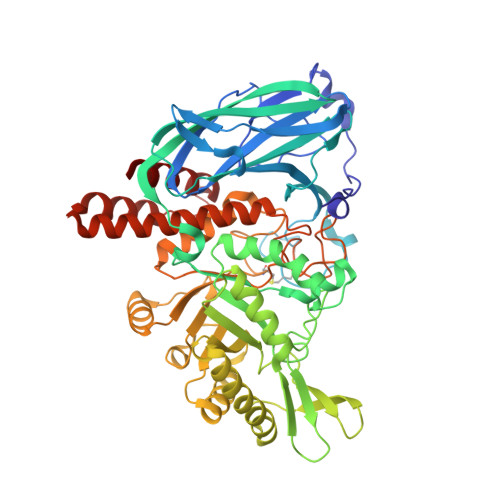
Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Roth, C., Petricevic, M., John, A., Goddard-Borger, E.D., Davies, G.J., Williams, S.J.(2016) Chem Commun (Camb) 52: 11096-11099
- PubMed: 27546776
- DOI: https://doi.org/10.1039/c6cc04649e
- Primary Citation of Related Structures:
5L7R, 5L7U, 5L7V - PubMed Abstract:
Bacteroides vulgatus is a member of the human microbiota whose abundance is increased in patients with Crohn's disease. We show that a B. vulgatus glycoside hydrolase from the carbohydrate active enzyme family GH123, BvGH123, is an N-acetyl-β-galactosaminidase that acts with retention of stereochemistry, and, through a 3-D structure in complex with Gal-thiazoline, provide evidence in support of a neighbouring group participation mechanism.
Organizational Affiliation:
Department of Chemistry, University of York, Heslington, York, UK. gideon.davies@york.ac.uk.