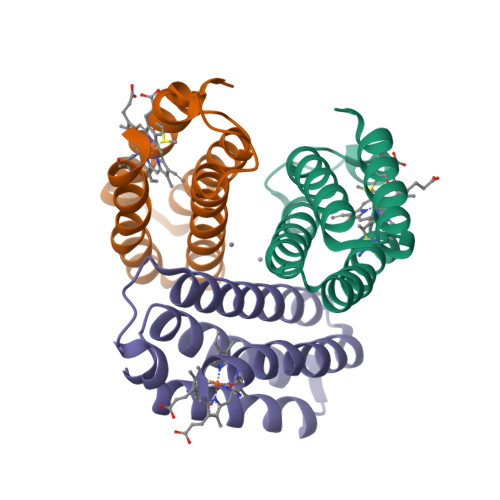
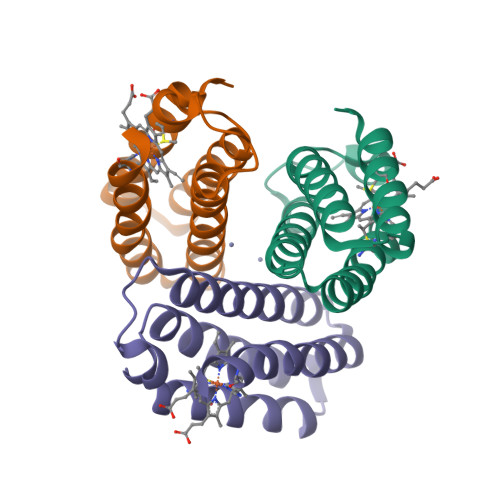
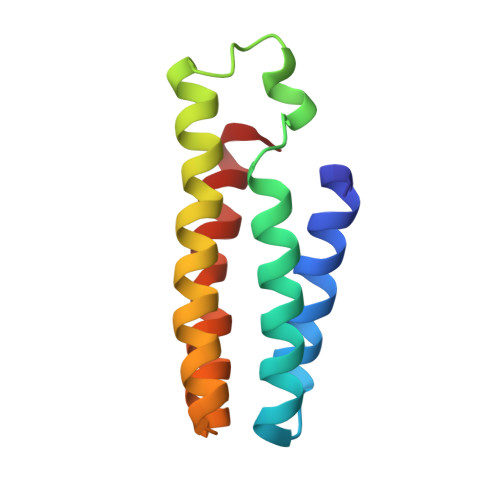
Structural characterization of a microperoxidase inside a metal-directed protein cage.
Ni, T.W., Tezcan, F.A.(2010) Angew Chem Int Ed Engl 49: 7014-7018
- PubMed: 20721993
- DOI: https://doi.org/10.1002/anie.201001487
- Primary Citation of Related Structures:
3M15, 3M4B, 3M4C
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, MC 0356, La Jolla, CA 92093, USA.