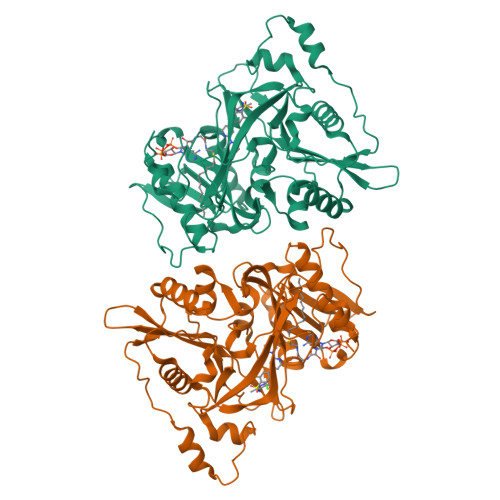
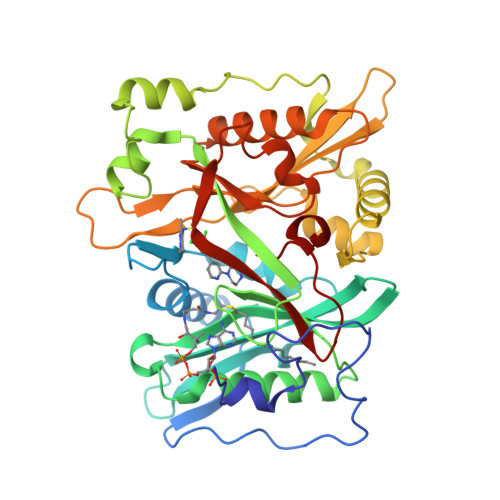
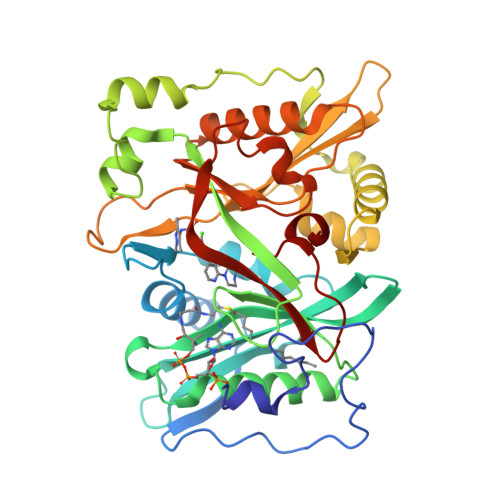
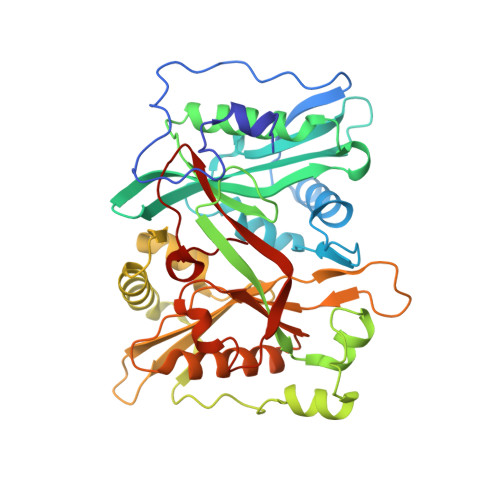
Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
Qiu, W., Hutchinson, A., Wernimont, A., Lin, Y.-H., Kania, A., Ravichandran, M., Kozieradzki, I., Cossar, D., Schapira, M., Arrowsmith, C.H., Bountra, C., Weigelt, J., Edwards, A.M., Wyatt, P.G., Ferguson, M.A.J., Frearson, J.A., Brand, S.Y., Robinson, D.A., Bochkarev, A., Hui, R.To be published.