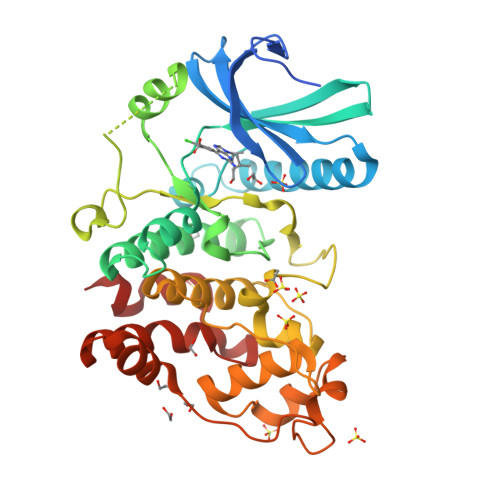
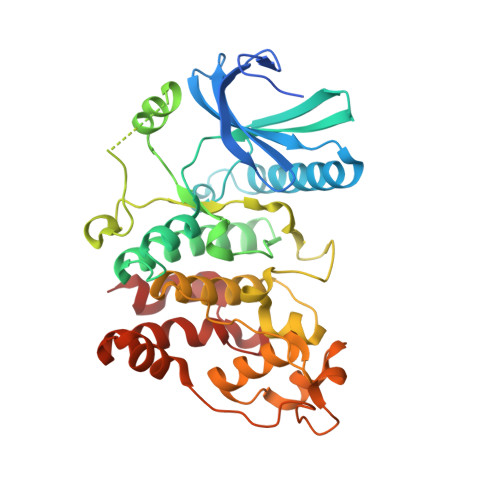
Structure of Human Serine-Arginine-Rich Protein- Specific Kinase 2 (Srpk2) Bound to Purvalanol B
Pike, A.C.W., Savitsky, P., Fedorov, O., Krojer, T., Ugochukwu, E., von Delft, F., Gileadi, O., Edwards, A., Arrowsmith, C.H., Weigelt, J., Bountra, C., Knapp, S.To be published.