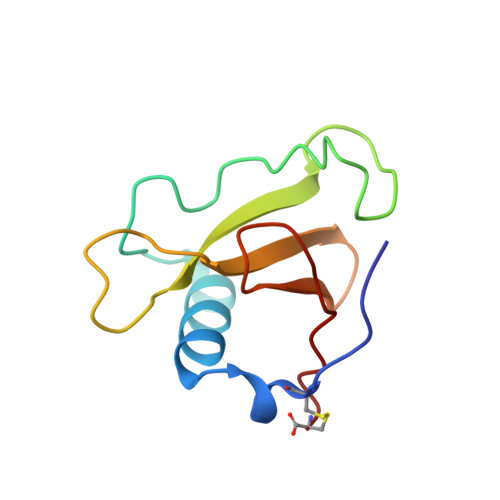
Determination and restrained least-squares refinement of the structures of ribonuclease Sa and its complex with 3'-guanylic acid at 1.8 A resolution.
Sevcik, J., Dodson, E.J., Dodson, G.G.(1991) Acta Crystallogr B 47: 240-253
- PubMed: 1654932
- Primary Citation of Related Structures:
1SAR, 2SAR - PubMed Abstract:
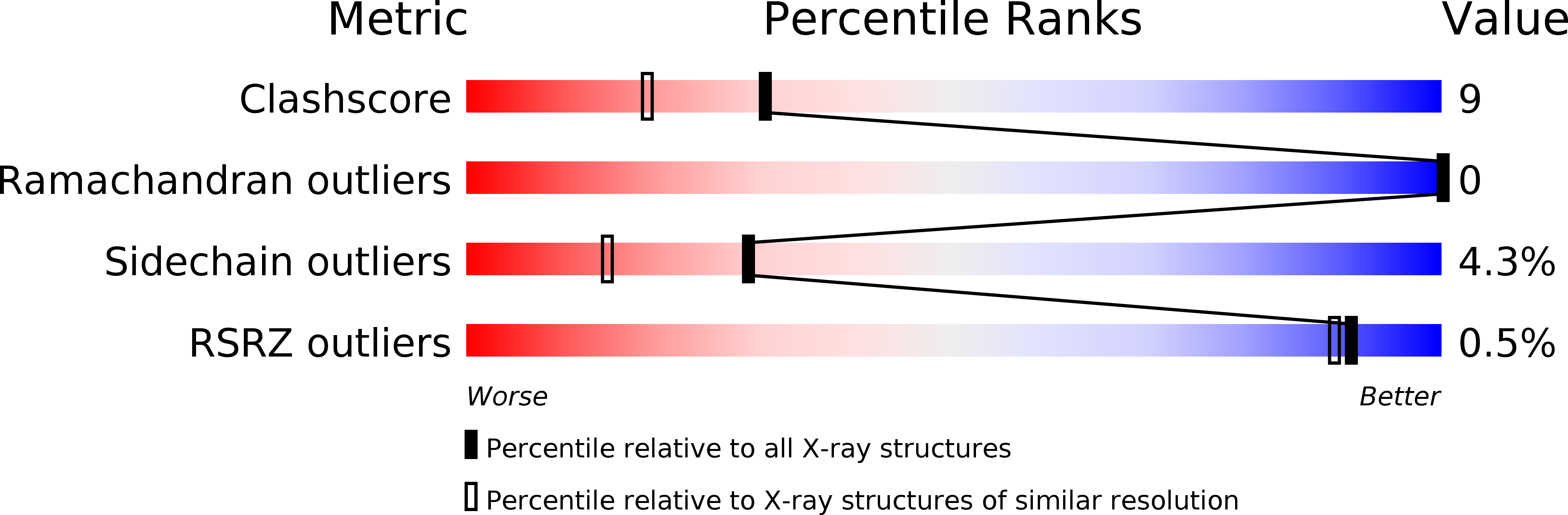
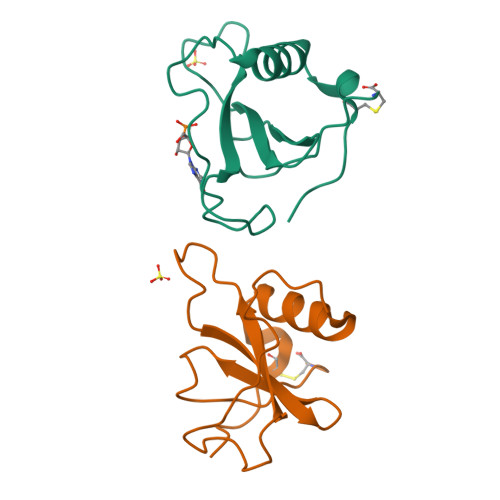
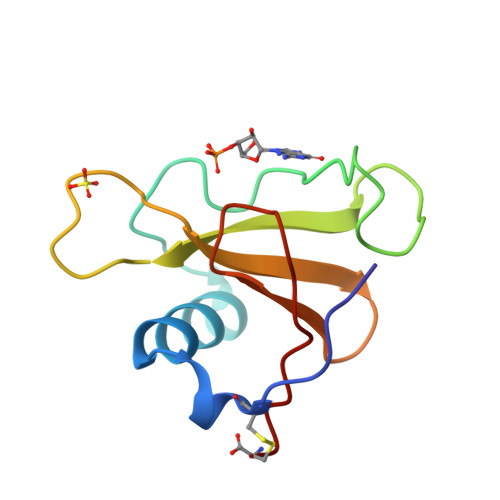
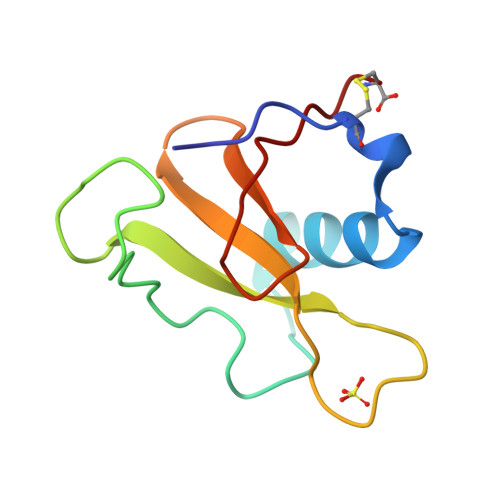
The crystal structures of ribonuclease from Streptomyces aureofaciens (RNase Sa) and its complex with 3'-guanylic acid (guanosine 3'-monophosphate, 3'-GMP) have been determined by the method of isomorphous replacement. The atomic parameters have been refined by restrained least-squares minimization using data in the resolution range 10.0-1.8 A. All protein atoms and more than 230 water atoms in the two crystal structures have been refined to crystallographic R factors of 0.172 and 0.175 respectively. The estimated r.m.s. error in the atomic positions ranges from 0.2 A for well-defined atoms to about 0.5 A for more poorly defined atoms. There are two enzyme molecules in the asymmetric unit, built independently, and referred to as molecules A and B. The value of the average B factor for protein atoms in both structures is about 19 A2 and for water molecules about 35 A2. Electron density for the substrate analogue 3'-GMP was found only at the active site of molecule A. The density was very clear and the positions of all 3'-GMP atoms were refined with precision comparable to that of the protein.
Organizational Affiliation:
Institute of Molecular Biology, Slovak Academy of Sciences, Bratislava, Czechoslovakia.