Structural basis of RNA-dependent recruitment of glutamine to the genetic code
Oshikane, H., Sheppard, K., Fukai, S., Nakamura, Y., Ishitani, R., Numata, T., Sherrer, R.L., Feng, L., Schmitt, E., Panvert, M., Blanquet, S., Mechulam, Y., Soll, D., Nureki, O.(2006) Science 312: 1950-1954
- PubMed: 16809540
- DOI: https://doi.org/10.1126/science.1128470
- Primary Citation of Related Structures:
2D6F - PubMed Abstract:
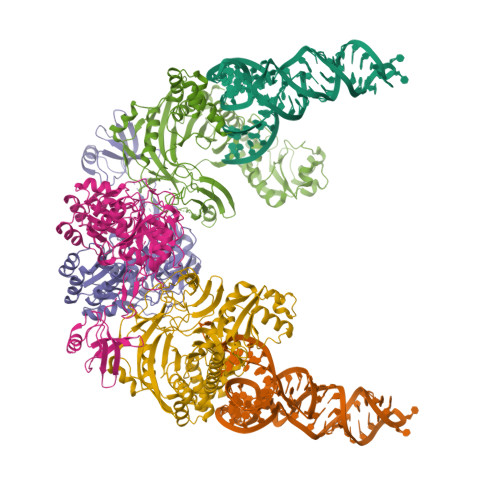
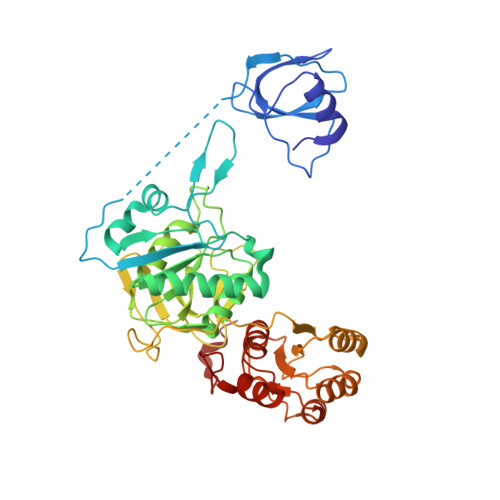
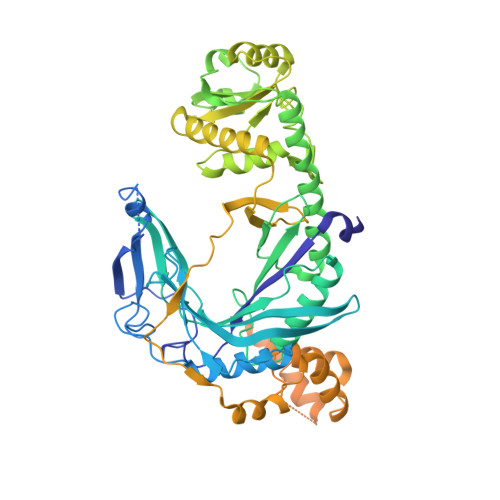
Glutaminyl-transfer RNA (Gln-tRNA(Gln)) in archaea is synthesized in a pretranslational amidation of misacylated Glu-tRNA(Gln) by the heterodimeric Glu-tRNA(Gln) amidotransferase GatDE. Here we report the crystal structure of the Methanothermobacter thermautotrophicus GatDE complexed to tRNA(Gln) at 3.15 angstroms resolution. Biochemical analysis of GatDE and of tRNA(Gln) mutants characterized the catalytic centers for the enzyme's three reactions (glutaminase, kinase, and amidotransferase activity). A 40 angstrom-long channel for ammonia transport connects the active sites in GatD and GatE. tRNA(Gln) recognition by indirect readout based on shape complementarity of the D loop suggests an early anticodon-independent RNA-based mechanism for adding glutamine to the genetic code.
Organizational Affiliation:
Department of Biological Information, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi, Kanagawa 226-8501, Japan.